1764 results
X-ray diffraction data for the CRYSTAL STRUCTURE OF A DTDP-4-DEHYDRORHAMNOSE REDUCTASE, RFBD ORTHOLOG (CA_C2315) FROM CLOSTRIDIUM ACETOBUTYLICUM ATCC 824 AT 2.05 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.17/0.21
Resolution: 2.05 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of Formyltetrahydrofolate deformylase (YP_105254.1) from BURKHOLDERIA MALLEI ATCC 23344 at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.19/0.22
Resolution: 1.90 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of phage related exonuclease (YP_719632.1) from HAEMOPHILUS SOMNUS 129PT at 2.15 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.17/0.21
Resolution: 2.15 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of Histidinol-phosphate aminotransferase (EC 2.6.1.9) (Imidazole acetol-phosphate transferase) (tm1040) from Thermotoga maritima at 2.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.18/0.24
Resolution: 2.40 Å
R/Rfree: 0.18/0.24
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE HD SUPERFAMILY HYDROLASE (BH1327) FROM BACILLUS HALODURANS AT 1.90 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.17/0.22
Resolution: 1.90 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF A XISI-LIKE PROTEIN (AVA_3825) FROM ANABAENA VARIABILIS ATCC 29413 AT 1.30 A RESOLUTION


First author:
W.C. Hwang
Resolution: 1.30 Å
R/Rfree: 0.18/0.20
Resolution: 1.30 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the CRYSTAL STRUCTURE OF a DUF2131 family protein (SAMA_2911) FROM SHEWANELLA AMAZONENSIS SB2B AT 2.50 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.21/0.25
Resolution: 2.50 Å
R/Rfree: 0.21/0.25
X-ray diffraction data for the Crystal structure of a putative thioesterase (pmt_2055) from prochlorococcus marinus str. mit 9313 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.16/0.18
Resolution: 1.50 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE CARBOXYLESTERASE (LP_2923) FROM LACTOBACILLUS PLANTARUM WCFS1 AT 1.70 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.17/0.20
Resolution: 1.70 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a putative yfit-like metal-dependent hydrolase (bh0186) from bacillus halodurans c-125 at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.15/0.20
Resolution: 2.30 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the 1.7 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii with Tyrosine of Tag Bound to Active Site

First author:
G. Minasov
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.8 A resolution


First author:
M. Ma
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD


X-ray diffraction data for the 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis


X-ray diffraction data for the 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae


X-ray diffraction data for the 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.

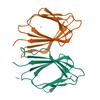
X-ray diffraction data for the 2.2 Angstrom Resolution Crystal Structure of P-Hydroxybenzoate Hydroxylase from Pseudomonas putida in Complex with FAD.


First author:
L.Shuvalova G.Minasov
Gene name: pobA
Resolution: 2.20 Å
R/Rfree: 0.17/0.22
Gene name: pobA
Resolution: 2.20 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the 2.0 Angstrom Resolution Crystal Structure of Transaldolase B (TalA) from Francisella tularensis in Covalent Complex with Fructose 6-Phosphate


X-ray diffraction data for the 1.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus (IDP00699) in complex with NAD+


X-ray diffraction data for the 2.50 angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with phosphoenolpyruvate


X-ray diffraction data for the 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica.


X-ray diffraction data for the 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD.


X-ray diffraction data for the 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium.


X-ray diffraction data for the The X-ray crystal structure of Streptococcus pneumoniae Fatty Acid Kinase (Fak) B1 protein loaded with myristic acid (C14:0) to 1.69 Angstrom resolution


First author:
M.G. Cuypers
Resolution: 1.69 Å
R/Rfree: 0.15/0.20
Resolution: 1.69 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the Crystal structure of a Putative phosphoribosylformylglycinamidine cyclo-ligase (BDI_2101) from Parabacteroides distasonis ATCC 8503 at 1.91 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.91 Å
R/Rfree: 0.15/0.19
Resolution: 1.91 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a biotin protein ligase-like protein of unknown function (tm1040_0394) from silicibacter sp. tm1040 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.18/0.23
Resolution: 1.70 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PROTEIN WITH UNKNOWN FUNCTION FROM DUF3598 FAMILY (NPUN_R4044) FROM NOSTOC PUNCTIFORME PCC 73102 AT 1.75 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.17/0.21
Resolution: 1.75 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.40 A resolution (PMSF inhibitor complex structure)


First author:
M. Levisson
Resolution: 2.40 Å
R/Rfree: 0.16/0.21
Resolution: 2.40 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of a binding protein component of ABC phosphonate transporter (PA3383) from Pseudomonas aeruginosa at 1.97 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.97 Å
R/Rfree: 0.16/0.19
Resolution: 1.97 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE THIOESTERASE (SSO2295) FROM SULFOLOBUS SOLFATARICUS AT 1.91 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.91 Å
R/Rfree: 0.22/0.26
Resolution: 1.91 Å
R/Rfree: 0.22/0.26
X-ray diffraction data for the Crystal structure of an osmc-like hydroperoxide resistance protein (jann_2040) from jannaschia sp. ccs1 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.14/0.16
Resolution: 1.70 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of Fumarase of FUM-1 (NP_069927.1) from Archaeoglobus Fulgidus at 1.66 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.66 Å
R/Rfree: 0.17/0.20
Resolution: 1.66 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PROTEIN WITH UNKNOWN FUNCTION FROM DUF3224 FAMILY (SO_1590) FROM SHEWANELLA ONEIDENSIS MR-1 AT 1.84 A RESOLUTION

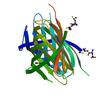
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.84 Å
R/Rfree: 0.18/0.21
Resolution: 1.84 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE ANTIBIOTIC BIOSYNTHESIS MONOOXYGENASE (CC_2132) FROM CAULOBACTER CRESCENTUS CB15 AT 1.35 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.35 Å
R/Rfree: 0.19/0.21
Resolution: 1.35 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the Crystal structure of a n-myc downstream regulated 2 protein (ndrg2, syld, ndr2, ai182517, au040374) from mus musculus at 1.70 A resolution


First author:
J. Hwang
Resolution: 1.70 Å
R/Rfree: 0.15/0.18
Resolution: 1.70 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of carbamoylphosphate synthase large subunit (split gene in MJ) (ZP_00538348.1) from Exiguobacterium sp. 255-15 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a two domain protein with unknown function (bt_3535) from bacteroides thetaiotaomicron vpi-5482 at 2.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.60 Å
R/Rfree: 0.20/0.24
Resolution: 2.60 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of Putative SnoaL-like polyketide cyclase (YP_563807.1) from SHEWANELLA DENITRIFICANS OS-217 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.19/0.24
Resolution: 1.70 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of protein of unknown function with cystatin-like fold (NP_639274.1) from Xanthomonas campestris at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.17/0.20
Resolution: 1.90 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a putative polyketide cyclase (lferr_0659) from acidithiobacillus ferrooxidans atcc at 1.76 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.76 Å
R/Rfree: 0.17/0.21
Resolution: 1.76 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a pilus assembly protein CpaE (CC_2943) from Caulobacter crescentus CB15 at 1.75 A resolution (PSI Community Target, Shapiro)


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.18/0.21
Resolution: 1.75 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a nitroreductase-like family protein (pnba, bh06130) from bartonella henselae str. houston-1 at 1.45 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.45 Å
R/Rfree: 0.15/0.17
Resolution: 1.45 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of a Putative Pii-Like Signaling Protein (YP_323533.1) from ANABAENA VARIABILIS ATCC 29413 at 2.35 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.35 Å
R/Rfree: 0.22/0.27
Resolution: 2.35 Å
R/Rfree: 0.22/0.27
X-ray diffraction data for the Crystal structure of a SusD-like protein (BF3747) from Bacteroides fragilis NCTC 9343 at 2.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.70 Å
R/Rfree: 0.18/0.21
Resolution: 2.70 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a CheY-like protein (tadZ) from Pseudomonas aeruginosa PAO1 at 2.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.20/0.23
Resolution: 2.40 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Crystal structure of a putative metal-dependent phosphoesterase (BAD_1165) from bifidobacterium adolescentis atcc 15703 at 1.94 A resolution


First author:
G.W. Han
Resolution: 1.94 Å
R/Rfree: 0.16/0.19
Resolution: 1.94 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE ANTIBIOTIC BIOSYNTHESIS MONOOXYGENASE (SPO2313) FROM SILICIBACTER POMEROYI DSS-3 AT 1.30 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.13/0.15
Resolution: 1.30 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Crystal structure of protein of unknown function with ferritin-like fold (YP_832262.1) from Arthrobacter sp. FB24 at 2.33 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.33 Å
R/Rfree: 0.21/0.26
Resolution: 2.33 Å
R/Rfree: 0.21/0.26
X-ray diffraction data for the Crystal structure of an alpha/beta hydrolase (YP_496220.1) from Novosphingobium aromaticivorans DSM 12444 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.16/0.20
Resolution: 1.50 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of a duf574 family protein (sav_2177) from streptomyces avermitilis ma-4680 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.20/0.25
Resolution: 1.80 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of 30S ribosomal protein S6 (TM0603) from Thermotoga maritima at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.18/0.24
Resolution: 1.70 Å
R/Rfree: 0.18/0.24
X-ray diffraction data for the Crystal structure of a putative two-domain sugar hydrolase (BACCAC_02064) from Bacteroides caccae ATCC 43185 at 1.35 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.35 Å
R/Rfree: 0.11/0.13
Resolution: 1.35 Å
R/Rfree: 0.11/0.13
X-ray diffraction data for the Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target)


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.14/0.17
Resolution: 1.90 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of a glycosyl hydrolase (BACOVA_03624) from Bacteroides ovatus at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.18/0.22
Resolution: 2.30 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a putative adhesin (BACOVA_04077) from Bacteroides ovatus at 2.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.21/0.24
Resolution: 2.50 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of a TetR family transcriptional regulator (Caur_2714) from CHLOROFLEXUS AURANTIACUS J-10-FL at 2.56 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.56 Å
R/Rfree: 0.21/0.24
Resolution: 2.56 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the CRYSTAL STRUCTURE OF AN UNCHARACTERIZED PROTEIN WITH A CYSTATIN-LIKE FOLD (CC_2572) FROM CAULOBACTER VIBRIOIDES AT 1.40 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.14/0.15
Resolution: 1.40 Å
R/Rfree: 0.14/0.15
X-ray diffraction data for the Crystal structure of an uncharacterized protein (PARMER_03598) from Parabacteroides merdae ATCC 43184 at 1.50 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of a putative cell surface protein (BACOVA_01565) from Bacteroides ovatus ATCC 8483 at 2.05 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.18/0.22
Resolution: 2.05 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a putative hydrolase (lpg1103) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 at 1.15 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.15 Å
R/Rfree: 0.14/0.16
Resolution: 1.15 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of putative MarR-like transcription regulator (NP_978771.1) from Bacillus cereus ATCC 10987 at 2.38 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.38 Å
R/Rfree: 0.23/0.25
Resolution: 2.38 Å
R/Rfree: 0.23/0.25
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE SUGAR PHOSPHATE ISOMERASE/EPIMERASE (AVA4194) FROM ANABAENA VARIABILIS ATCC 29413 AT 1.78 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.78 Å
R/Rfree: 0.18/0.22
Resolution: 1.78 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a duf1048 protein with a left-handed superhelix fold (bce_3448) from bacillus cereus atcc 10987 at 2.05 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.18/0.23
Resolution: 2.05 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of SusD-like carbohydrate binding protein (YP_001298396.1) from Bacteroides vulgatus ATCC 8482 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.15/0.17
Resolution: 1.70 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the CRYSTAL STRUCTURE OF A DIMERIC FERREDOXIN-LIKE PROTEIN (JCVI_PEP_1096672785533) FROM UNCULTURED MARINE ORGANISM AT 2.20 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.23/0.29
Resolution: 2.20 Å
R/Rfree: 0.23/0.29
X-ray diffraction data for the Crystal structure of a cupin-like protein (tm1010) from thermotoga maritima msb8 at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.16/0.22
Resolution: 1.90 Å
R/Rfree: 0.16/0.22
X-ray diffraction data for the Crystal structure of a putative periplasmic protein of unknown function (bvu_2443) from bacteroides vulgatus atcc 8482 at 1.64 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.64 Å
R/Rfree: 0.17/0.21
Resolution: 1.64 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the CRYSTAL STRUCTURE OF a putative oxidoreductase of the DsrE/DsrF-like family (SSO1126) FROM SULFOLOBUS SOLFATARICUS P2 AT 2.09 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.09 Å
R/Rfree: 0.18/0.20
Resolution: 2.09 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of a putative inositol catabolism protein iole (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE ACETYLACETONE DIOXYGENASE (MPE_A3659) FROM METHYLIBIUM PETROLEIPHILUM PM1 AT 1.50 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.18/0.21
Resolution: 1.50 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a duf89 family protein (ph1575) from pyrococcus horikoshii at 2.04 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.04 Å
R/Rfree: 0.14/0.18
Resolution: 2.04 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.12 A resolution (paraoxon inhibitor complex structure)


First author:
M. Levisson
Resolution: 2.12 Å
R/Rfree: 0.17/0.21
Resolution: 2.12 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of Gamma-glutamyl phosphate reductase (yor323c) from Saccharomyces cerevisiae at 2.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.29 Å
R/Rfree: 0.21/0.25
Resolution: 2.29 Å
R/Rfree: 0.21/0.25
X-ray diffraction data for the Crystal structure of Putative sugar binding protein (YP_001300177.1) from Bacteroides vulgatus ATCC 8482 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a putative 3-keto-5-aminohexanoate cleavage enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.72 Å
R/Rfree: 0.15/0.18
Resolution: 1.72 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative 4-methylmuconolactone methylisomerase (YP_295714.1) from Ralstonia eutropha JMP134 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.17/0.20
Resolution: 2.00 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a putative dipeptidyl-peptidase VI (BT_1314) from Bacteroides thetaiotaomicron VPI-5482 at 1.75 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.15/0.18
Resolution: 1.75 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE DEHYDRATASE FROM THE NTF2-LIKE FAMILY (SAV_4671) FROM STREPTOMYCES AVERMITILIS AT 2.10 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.20/0.24
Resolution: 2.10 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile.

First author:
G. Minasov
Resolution: 2.00 Å
R/Rfree: 0.21/0.27
Resolution: 2.00 Å
R/Rfree: 0.21/0.27
X-ray diffraction data for the 2.23 Angstrom resolution crystal structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase (murA) from Listeria monocytogenes EGD-e

First author:
A.S. Halavaty
Gene name: murA
Resolution: 2.23 Å
R/Rfree: 0.20/0.24
Gene name: murA
Resolution: 2.23 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the 1.8 Angstrom Resolution Crystal Structure of K135M Mutant of Transaldolase B (TalA) from Francisella tularensis in Complex with Fructose 6-phosphate

First author:
G. Minasov
Gene name: talA
Resolution: 1.80 Å
R/Rfree: 0.16/0.19
Gene name: talA
Resolution: 1.80 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the 1.8 Angstrom resolution crystal structure of dihydroorotase (pyrC) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2

First author:
G. Minasov
Gene name: pyrC
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
Gene name: pyrC
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the 1.18 Angstrom resolution crystal structure of uncharacterized protein lmo1340 from Listeria monocytogenes EGD-e

First author:
A.S. Halavaty
Gene name: -
Resolution: 1.18 Å
R/Rfree: 0.15/0.18
Gene name: -
Resolution: 1.18 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative 3-oxoacyl-(acyl carrier protein) reductase from Bacillus anthracis at 1.87 A resolution

First author:
J. Hou
Resolution: 1.87 Å
R/Rfree: 0.17/0.21
Resolution: 1.87 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the 1.75 Angstrom resolution crystal structure of a putative NTP pyrophosphohydrolase (yfaO) from Salmonella typhimurium LT2

First author:
A.S. Halavaty
Gene name: yfaO
Resolution: 1.75 Å
R/Rfree: 0.19/0.22
Gene name: yfaO
Resolution: 1.75 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the 2.60 Angstrom resolution crystal structure of putative ribose 5-phosphate isomerase from Toxoplasma gondii ME49 in complex with DL-Malic acid

First author:
A.S. Halavaty
Resolution: 2.60 Å
R/Rfree: 0.16/0.20
Resolution: 2.60 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the 1.90 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus in complex with NAD+ and BME-free Cys289

First author:
A.S. Halavaty
Gene name: betB
Resolution: 1.90 Å
R/Rfree: 0.14/0.19
Gene name: betB
Resolution: 1.90 Å
R/Rfree: 0.14/0.19
X-ray diffraction data for the Crystal structure of putative dioxygenase (YP_555069.1) from Burkholderia xenovorans LB400 at 1.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.14/0.17
Resolution: 1.40 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of uncharacterized conserved protein with double-stranded beta-helix domain (YP_001338853.1) from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.15/0.20
Resolution: 1.80 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the 2.2 Angstrom Resolution Crystal Structure of putative UDP-N-acetylglucosamine 2-epimerase from Listeria monocytogenes

First author:
G. Minasov
Resolution: 2.20 Å
R/Rfree: 0.19/0.23
Resolution: 2.20 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the 2.35 Angstrom resolution crystal structure of a putative tRNA (guanine-7-)-methyltransferase (trmD) from Staphylococcus aureus subsp. aureus MRSA252

First author:
A.S. Halavaty
Gene name: trmD
Resolution: 2.35 Å
R/Rfree: 0.21/0.26
Gene name: trmD
Resolution: 2.35 Å
R/Rfree: 0.21/0.26
X-ray diffraction data for the 2.6 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) in complex with
NAD+ and BME-free Cys289

First author:
G.Minasov A.S.Halavaty
Gene name: betB
Resolution: 2.60 Å
R/Rfree: 0.19/0.22
Gene name: betB
Resolution: 2.60 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the 2.49 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate

First author:
A.S. Halavaty
Gene name: aroE
Resolution: 2.49 Å
R/Rfree: 0.19/0.25
Gene name: aroE
Resolution: 2.49 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile.

First author:
G. Minasov
Resolution: 1.63 Å
R/Rfree: 0.16/0.19
Resolution: 1.63 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the 1.55 Angstrom Resolution Crystal Structure of Peptidase T (pepT-1) from Bacillus anthracis str. 'Ames Ancestor'.

First author:
G. Minasov
Gene name: pepT-1
Resolution: 1.55 Å
R/Rfree: 0.15/0.17
Gene name: pepT-1
Resolution: 1.55 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the 1.85 Angstrom Resolution Crystal Structure of Transaldolase B (talA) from Francisella tularensis.

First author:
G. Minasov
Gene name: talA
Resolution: 1.85 Å
R/Rfree: 0.15/0.19
Gene name: talA
Resolution: 1.85 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the 2.25 Angstrom resolution crystal structure of UDP-N-acetylmuramate--L-alanine ligase (murC) from Yersinia pestis CO92 in complex with AMP

First author:
A.S. Halavaty
Gene name: murC
Resolution: 2.25 Å
R/Rfree: 0.18/0.22
Gene name: murC
Resolution: 2.25 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the 2.00 Angstrom resolution crystal structure of a catalytic subunit of an aspartate carbamoyltransferase (pyrB) from Yersinia pestis CO92

First author:
A.S. Halavaty
Gene name: pyrB
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
Gene name: pyrB
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the 2.20 Angstrom resolution crystal structure of protein YE0340 of unidentified function from Yersinia enterocolitica subsp. enterocolitica 8081]

First author:
A.S. Halavaty
Resolution: 2.20 Å
R/Rfree: 0.23/0.29
Resolution: 2.20 Å
R/Rfree: 0.23/0.29
X-ray diffraction data for the 2.30 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-free Cys289

First author:
A.S. Halavaty
Gene name: betB
Resolution: 2.30 Å
R/Rfree: 0.16/0.22
Gene name: betB
Resolution: 2.30 Å
R/Rfree: 0.16/0.22