6726 results
X-ray diffraction data for the Crystal structure of a putative conserved lipoprotein (NT01CX_1156) from Clostridium novyi NT at 2.70 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.70 Å
R/Rfree: 0.20/0.20
Resolution: 2.70 Å
R/Rfree: 0.20/0.20
X-ray diffraction data for the Crystal structure of a DUF1672 family protein (SAV1486) from Staphylococcus aureus subsp. aureus Mu50 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.18/0.22
Resolution: 1.80 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a superantigen-like protein (SAV0433) from Staphylococcus aureus MU50 at 2.05 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.19/0.24
Resolution: 2.05 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of an exotoxin 6 (SAV0422) from Staphylococcus aureus subsp. aureus Mu50 at 2.25 A resolution

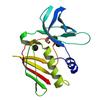
First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.25 Å
R/Rfree: 0.19/0.24
Resolution: 2.25 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a putative peptide binding protein (RUMGNA_00914) from Ruminococcus gnavus ATCC 29149 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.16/0.20
Resolution: 1.60 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of a putative metal binding protein RUMGNA_00854 (ZP_02040092.1) from Ruminococcus gnavus ATCC 29149 at 1.30 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.14/0.19
Resolution: 1.30 Å
R/Rfree: 0.14/0.19
X-ray diffraction data for the Crystal structure of a DUF3887 family protein (RUMGNA_01855) from Ruminococcus gnavus ATCC 29149 at 2.25 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.25 Å
R/Rfree: 0.19/0.21
Resolution: 2.25 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the Crystal structure of a NTF2-like protein (EUBSIR_01394) from Eubacterium siraeum DSM 15702 at 2.56 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.56 Å
R/Rfree: 0.25/0.30
Resolution: 2.56 Å
R/Rfree: 0.25/0.30
X-ray diffraction data for the Crystal structure of a protein with unknown function (CLOLEP_02462) from Clostridium leptum DSM 753 at 2.10 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.16/0.20
Resolution: 2.10 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of a DUF5037 family protein (RUMGNA_01148) from Ruminococcus gnavus ATCC 29149 at 2.55 A resolution


First author:
Joint center for structural genomics (JCSG)
Resolution: 2.55 Å
R/Rfree: 0.19/0.22
Resolution: 2.55 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a putative transcription regulator (EUBSIR_01389) from Eubacterium siraeum DSM 15702 at 2.15 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.20/0.25
Resolution: 2.15 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of a DHHW family protein (EUBSIR_00411) from Eubacterium siraeum DSM 15702 at 1.49 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.49 Å
R/Rfree: 0.13/0.16
Resolution: 1.49 Å
R/Rfree: 0.13/0.16
X-ray diffraction data for the Crystal structure of an apag protein (PA1934) from pseudomonas aeruginosa pao1 at 1.83 A resolution

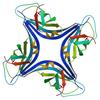
First author:
Joint center for structural genomics (JCSG)
Resolution: 1.83 Å
R/Rfree: 0.19/0.22
Resolution: 1.83 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of an apag protein (PA1934) from pseudomonas aeruginosa pao1 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.18/0.22
Resolution: 1.50 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of an immunoglobulin-like protein (PA1606) from PSEUDOMONAS AERUGINOSA at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.18/0.21
Resolution: 1.50 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a putative secreted protein (PA3611) from PSEUDOMONAS AERUGINOSA at 1.60 A resolution


First author:
D. Das
Resolution: 1.60 Å
R/Rfree: 0.16/0.21
Resolution: 1.60 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of a Pfam DUF1093 family protein (BSU39620) from Bacillus subtilis at 2.15 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.18/0.22
Resolution: 2.15 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a DUF1541 family protein (ydhK) from Bacillus subtilis subsp. subtilis str. 168 at 2.00 A resolution

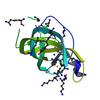
First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.18/0.26
Resolution: 2.00 Å
R/Rfree: 0.18/0.26
X-ray diffraction data for the Crystal structure of a putative hydrolase (BT3161) from Bacteroides thetaiotaomicron VPI-5482 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.15/0.19
Resolution: 1.80 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a yobA protein (BSU18810) from Bacillus subtilis subsp. subtilis str. 168 at 1.55 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.55 Å
R/Rfree: 0.19/0.22
Resolution: 1.55 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a DUF4309 family protein (YjgB) from Bacillus subtilis subsp. subtilis str. 168 at 2.13 A resolution

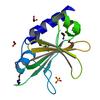
First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.13 Å
R/Rfree: 0.19/0.23
Resolution: 2.13 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of a putative lipoprotein (ycdA) from Bacillus subtilis subsp. subtilis str. 168 at 2.62 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.62 Å
R/Rfree: 0.21/0.24
Resolution: 2.62 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of an uncharacterized protein (yncM) from Bacillus subtilis subsp. subtilis str. 168 at 1.55 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.55 Å
R/Rfree: 0.19/0.22
Resolution: 1.55 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a DUF4853 family protein (ACTODO_00621) from Actinomyces odontolyticus ATCC 17982 at 2.65 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.65 Å
R/Rfree: 0.23/0.26
Resolution: 2.65 Å
R/Rfree: 0.23/0.26
X-ray diffraction data for the Crystal structure of a sugar ABC transporter (ACTODO_00688) from Actinomyces odontolyticus ATCC 17982 at 2.60 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.60 Å
R/Rfree: 0.21/0.22
Resolution: 2.60 Å
R/Rfree: 0.21/0.22
X-ray diffraction data for the Crystal structure of an atmB (putative membrane lipoprotein) from Streptococcus mutans UA159 at 1.91 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.91 Å
R/Rfree: 0.18/0.23
Resolution: 1.91 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of a putative lipoprotein (RUMGNA_00858) from Ruminococcus gnavus ATCC 29149 at 1.76 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.76 Å
R/Rfree: 0.16/0.18
Resolution: 1.76 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of a putative methionine-binding lipoprotein (BSU32730) from Bacillus subtilis subsp. subtilis str. 168 at 1.95 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.17/0.23
Resolution: 1.95 Å
R/Rfree: 0.17/0.23
X-ray diffraction data for the Crystal structure of a putative lipoprotein (CD630_1653) from Clostridium difficile 630 at 2.20 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.20/0.24
Resolution: 2.20 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of a putative TonB-dependent receptor (PA5505) from Pseudomonas aeruginosa PAO1 at 1.60 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.15/0.19
Resolution: 1.60 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a pheromone cOB1 precursor/lipoprotein, YaeC family (EF2496) from Enterococcus faecalis V583 at 2.10 A resolution


First author:
Joint center for structural genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.19/0.23
Resolution: 2.10 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of a pheromone cOB1 precursor/lipoprotein, YaeC family (EF2496) from Enterococcus faecalis V583 at 1.90 A resolution


First author:
Joint center for structural genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.19/0.24
Resolution: 1.90 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a lipoprotein, YaeC family (EF3198) from Enterococcus faecalis V583 at 1.80 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.18/0.21
Resolution: 1.80 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a putative lipoprotein (ACTODO_00931) from Actinomyces odontolyticus ATCC 17982 at 2.35 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.35 Å
R/Rfree: 0.16/0.21
Resolution: 2.35 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of a DUF4623 family protein (BACEGG_03550) from Bacteroides eggerthii DSM 20697 at 1.91 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.91 Å
R/Rfree: 0.15/0.19
Resolution: 1.91 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a streptavidin-like protein (BACEGG_01519) from Bacteroides eggerthii DSM 20697 at 1.25 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.25 Å
R/Rfree: 0.13/0.15
Resolution: 1.25 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Crystal structure of a DUF3571 family protein (ABAYE3784) from Acinetobacter baumannii AYE at 1.95 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.17/0.21
Resolution: 1.95 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a DUF4886 family protein (BACUNI_01406) from Bacteroides uniformis ATCC 8492 at 1.50 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.15/0.18
Resolution: 1.50 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative cell adhesion protein (BACEGG_00536) from Bacteroides eggerthii DSM 20697 at 1.67 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.67 Å
R/Rfree: 0.16/0.19
Resolution: 1.67 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a putative cell adhesion protein (BACOVA_04078) from Bacteroides ovatus ATCC 8483 at 2.80 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.80 Å
R/Rfree: 0.20/0.25
Resolution: 2.80 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of a beta-barrel like protein (ABAYE2633) from Acinetobacter baumannii AYE at 1.43 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.43 Å
R/Rfree: 0.12/0.15
Resolution: 1.43 Å
R/Rfree: 0.12/0.15
X-ray diffraction data for the Crystal structure of a DUF5043 family protein (BACUNI_01052) from Bacteroides uniformis ATCC 8492 at 1.85 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.16/0.18
Resolution: 1.85 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of a putative carbohydrate-binding protein (BACUNI_03838) from Bacteroides uniformis ATCC 8492 at 2.70 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.70 Å
R/Rfree: 0.21/0.24
Resolution: 2.70 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of a Putative thua-like protein (BACUNI_01602) from Bacteroides uniformis ATCC 8492 at 1.50 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.14/0.16
Resolution: 1.50 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of a Putative thua-like protein (BACUNI_01602) from Bacteroides uniformis ATCC 8492 at 2.30 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.16/0.21
Resolution: 2.30 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of a putative cell adhesion protein (BACOVA_04980) from Bacteroides ovatus ATCC 8483 at 2.18 A resolution

First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.18 Å
R/Rfree: 0.18/0.20
Resolution: 2.18 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of a fimbrial protein (BACOVA_04982) from Bacteroides ovatus ATCC 8483 at 1.85 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.16/0.18
Resolution: 1.85 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of a putative GDSL-like lipase (PARMER_00689) from Parabacteroides merdae ATCC 43184 at 2.86 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.86 Å
R/Rfree: 0.18/0.23
Resolution: 2.86 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of a porin-like protein (BACUNI_01323) from Bacteroides uniformis ATCC 8492 at 2.32 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.32 Å
R/Rfree: 0.17/0.19
Resolution: 2.32 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of a putative carbohydrate binding protein (BACUNI_04699) from Bacteroides uniformis ATCC 8492 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a DUF3823 family protein (BACUNI_03093) from Bacteroides uniformis ATCC 8492 at 1.90 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.18/0.21
Resolution: 1.90 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a putative cell adhesion protein (BACUNI_00621) from Bacteroides uniformis ATCC 8492 at 1.67 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.67 Å
R/Rfree: 0.19/0.22
Resolution: 1.67 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of an alginate lyase (BACOVA_01668) from Bacteroides ovatus at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.14/0.18
Resolution: 1.60 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the Crystal structure of an alginate lyase (BACOVA_01668) from Bacteroides ovatus at 1.95 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.16/0.20
Resolution: 1.95 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of a putative dipeptidyl aminopeptidase IV (BACOVA_01349) from Bacteroides ovatus ATCC 8483 at 2.48 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.48 Å
R/Rfree: 0.17/0.21
Resolution: 2.48 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a putative GDSL-like lipase (BACUNI_00748) from Bacteroides uniformis ATCC 8492 at 1.90 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.16/0.21
Resolution: 1.90 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of a putative GDSL-like lipase (BACOVA_04955) from Bacteroides ovatus ATCC 8483 at 2.10 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a sugar-binding protein (BACOVA_04391) from Bacteroides ovatus at 2.16 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.16 Å
R/Rfree: 0.20/0.23
Resolution: 2.16 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Crystal structure of a putative two-domain sugar hydrolase (BACCAC_02064) from Bacteroides caccae ATCC 43185 at 1.80 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.15/0.18
Resolution: 1.80 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative two-domain sugar hydrolase (BACCAC_02064) from Bacteroides caccae ATCC 43185 at 1.35 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.35 Å
R/Rfree: 0.11/0.13
Resolution: 1.35 Å
R/Rfree: 0.11/0.13
X-ray diffraction data for the Crystal structure of a putative protease (BACUNI_00178) from Bacteroides uniformis ATCC 8492 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.13/0.17
Resolution: 1.50 Å
R/Rfree: 0.13/0.17
X-ray diffraction data for the Crystal structure of a DUF1343 family protein (BF0371) from Bacteroides fragilis NCTC 9343 at 1.65 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.65 Å
R/Rfree: 0.15/0.18
Resolution: 1.65 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a DUF1343 family protein (BF0379) from Bacteroides fragilis NCTC 9343 at 1.30 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.12/0.15
Resolution: 1.30 Å
R/Rfree: 0.12/0.15
X-ray diffraction data for the Crystal structure of an exo-alpha-1,6-mannosidase (bacova_03347) from bacteroides ovatus at 1.60 a resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.13/0.17
Resolution: 1.60 Å
R/Rfree: 0.13/0.17
X-ray diffraction data for the Crystal structure of an exo-alpha-1,6-mannosidase (bacova_03626) from bacteroides ovatus at 1.70 a resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.13/0.17
Resolution: 1.70 Å
R/Rfree: 0.13/0.17
X-ray diffraction data for the Crystal structure of a putative member of duf 3244 protein family (BT_1867) from Bacteroides thetaiotaomicron VPI-5482 at 1.69 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.69 Å
R/Rfree: 0.18/0.21
Resolution: 1.69 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a leucine-rich repeat protein (BACEGG_03329) from Bacteroides eggerthii DSM 20697 at 2.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.70 Å
R/Rfree: 0.18/0.22
Resolution: 2.70 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a leucine-rich repeat protein (BT_0210) from Bacteroides thetaiotaomicron VPI-5482 at 2.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.80 Å
R/Rfree: 0.18/0.23
Resolution: 2.80 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of a STRUCTURAL GENOMICS, UNKNOWN FUNCTION (BACOVA_03430) from Bacteroides ovatus at 2.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.18/0.22
Resolution: 2.40 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of an endo-beta-N-acetylglucosaminidase (BT_3987) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.55 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.55 Å
R/Rfree: 0.14/0.16
Resolution: 1.55 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of a putative secreted protein (BF4250) from Bacteroides fragilis NCTC 9343 at 2.05 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.19/0.22
Resolution: 2.05 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a putative secreted protein (BF4250) from Bacteroides fragilis NCTC 9343 at 1.81 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.81 Å
R/Rfree: 0.17/0.20
Resolution: 1.81 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a cystatin-like protein (BACCAC_01506) from Bacteroides caccae ATCC 43185 at 1.69 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.69 Å
R/Rfree: 0.18/0.20
Resolution: 1.69 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of a Putative lipoprotein (BACOVA_00967) from Bacteroides ovatus at 2.08 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.08 Å
R/Rfree: 0.16/0.20
Resolution: 2.08 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.21/0.24
Resolution: 1.90 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target)


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.14/0.17
Resolution: 1.90 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of a putative protein binding protein (BACOVA_03105) from Bacteroides ovatus ATCC 8483 at 2.19 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.19 Å
R/Rfree: 0.22/0.24
Resolution: 2.19 Å
R/Rfree: 0.22/0.24
X-ray diffraction data for the Crystal structure of a putative extracellular heme-binding protein (DESPIG_02683) from Desulfovibrio piger ATCC 29098 at 1.24 A resolution

First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.24 Å
R/Rfree: 0.13/0.15
Resolution: 1.24 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Crystal structure of a probable hydrolytic enzyme (PA3053) from Pseudomonas aeruginosa PAO1 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.16/0.19
Resolution: 1.50 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a putative zinc-binding metallo-peptidase (BACCAC_01431) from Bacteroides caccae ATCC 43185 at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.09 Å
R/Rfree: 0.20/0.23
Resolution: 2.09 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Crystal structure of a DUF1312 family protein (RUMGNA_02503) from Ruminococcus gnavus ATCC 29149 at 2.20 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.18/0.21
Resolution: 2.20 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of an auxiliary nutrient binding protein (BT3507) from Bacteroides thetaiotaomicron VPI-5482 at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.18/0.23
Resolution: 2.30 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of a protein of unknown function (BACOVA_00267) from Bacteroides ovatus at 1.55 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.55 Å
R/Rfree: 0.17/0.19
Resolution: 1.55 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of a Multidomain protein including DUF1735 (BACOVA_03322) from Bacteroides ovatus at 2.80 A resolution


First author:
Joint center for structural genomics (jcsg)
Resolution: 2.80 Å
R/Rfree: 0.20/0.23
Resolution: 2.80 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Crystal structure of a STRUCTURAL GENOMICS, UNKNOWN FUNCTION (BACOVA_03322) from Bacteroides ovatus at 2.61 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.61 Å
R/Rfree: 0.20/0.24
Resolution: 2.61 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of a leucine-rich repeat protein (BT_1240) from Bacteroides thetaiotaomicron VPI-5482 at 1.99 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.99 Å
R/Rfree: 0.17/0.20
Resolution: 1.99 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a thioredoxin-like protein (BDI_1100) from Parabacteroides distasonis ATCC 8503 at 2.02 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.02 Å
R/Rfree: 0.18/0.21
Resolution: 2.02 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a putative flavoprotein (BACEGG_01620) from Bacteroides eggerthii DSM 20697 at 1.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.12/0.15
Resolution: 1.40 Å
R/Rfree: 0.12/0.15
X-ray diffraction data for the Crystal structure of a Putative flavoprotein (BACUNI_04544) from Bacteroides uniformis ATCC 8492 at 1.50 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of a DUF4784 family protein (BACCAC_01631) from Bacteroides caccae ATCC 43185 at 2.50 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.20/0.22
Resolution: 2.50 Å
R/Rfree: 0.20/0.22
X-ray diffraction data for the Crystal structure of a conjugative transposon lipoprotein (BACEGG_03088) from Bacteroides eggerthii DSM 20697 at 1.70 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.17/0.19
Resolution: 1.70 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of a DUF3836 family protein (BDI_3222) from Parabacteroides distasonis ATCC 8503 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of a putative neuraminidase (BACOVA_03493) from Bacteroides ovatus ATCC 8483 at 1.74 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.74 Å
R/Rfree: 0.17/0.21
Resolution: 1.74 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a putative glucoamylase (BACCAC_03554) from Bacteroides caccae ATCC 43185 at 2.05 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.18/0.22
Resolution: 2.05 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a putative glucoamylase (BACUNI_03963) from Bacteroides uniformis ATCC 8492 at 2.01 A resolution


First author:
Joint center for structural genomics (JCSG)
Resolution: 2.01 Å
R/Rfree: 0.14/0.17
Resolution: 2.01 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of a NigD-like immunity protein (BACCAC_03262) from Bacteroides caccae ATCC 43185 at 2.42 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.42 Å
R/Rfree: 0.18/0.21
Resolution: 2.42 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a putative immunoglobulin A1 protease (BACOVA_03286) from Bacteroides ovatus at 1.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.14/0.16
Resolution: 1.30 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of a protein with unknown function from DUF2233 family (BACOVA_00430) from Bacteroides ovatus at 1.80 A resolution


First author:
D. Das
Resolution: 1.80 Å
R/Rfree: 0.13/0.15
Resolution: 1.80 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Crystal structure of a putative alpha-L-fucosidase (BACOVA_04357) from Bacteroides ovatus ATCC 8483 at 1.59 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.59 Å
R/Rfree: 0.15/0.18
Resolution: 1.59 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative adhesin (BACOVA_04077) from Bacteroides ovatus at 2.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.21/0.24
Resolution: 2.50 Å
R/Rfree: 0.21/0.24