6726 results
X-ray diffraction data for the CRYSTAL STRUCTURE OF A DIMERIC FERREDOXIN-LIKE PROTEIN (CC_2267) FROM CAULOBACTER CRESCENTUS CB15 AT 1.64 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.64 Å
R/Rfree: 0.20/0.23
Resolution: 1.64 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Crystal structure of a dimeric ferredoxin-like protein (bb1511) from bordetella bronchiseptica rb50 at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.18/0.24
Resolution: 1.90 Å
R/Rfree: 0.18/0.24
X-ray diffraction data for the CRYSTAL STRUCTURE OF A DIMERIC FERREDOXIN-LIKE PROTEIN OF UNKNOWN FUNCTION (JANN_3925) FROM JANNASCHIA SP. CCS1 AT 2.30 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.18/0.22
Resolution: 2.30 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a Putative Pii-Like Signaling Protein (YP_323533.1) from ANABAENA VARIABILIS ATCC 29413 at 2.35 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.35 Å
R/Rfree: 0.22/0.27
Resolution: 2.35 Å
R/Rfree: 0.22/0.27
X-ray diffraction data for the CRYSTAL STRUCTURE OF A NTF-2 LIKE PROTEIN OF UNKNOWN FUNCTION (SO_0125) FROM SHEWANELLA ONEIDENSIS MR-1 AT 1.70 A RESOLUTION

First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.18/0.21
Resolution: 1.70 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a protein with unknown function (arth_0117) from arthrobacter sp. fb24 at 2.20 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.18/0.25
Resolution: 2.20 Å
R/Rfree: 0.18/0.25
X-ray diffraction data for the CRYSTAL STRUCTURE OF A NTF2-LIKE PROTEIN OF UNKNOWN FUNCTION (MFLA_0564) FROM METHYLOBACILLUS FLAGELLATUS KT AT 2.200 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.16/0.21
Resolution: 2.20 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of a ntf2-like protein of unknown function (sbal_0622) from shewanella baltica os155 at 1.75 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.15/0.19
Resolution: 1.75 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A NTF2-LIKE PROTEIN (CHU_1428) FROM CYTOPHAGA HUTCHINSONII ATCC 33406 AT 1.60 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.18/0.20
Resolution: 1.60 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of NTF2-like protein of unknown function in nutrient uptake (YP_427473.1) from RHODOSPIRILLUM RUBRUM ATCC 11170 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.17/0.22
Resolution: 1.70 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of ketosteroid isomerase-like protein (YP_049581.1) from ERWINIA CAROTOVORA ATROSEPTICA SCRI1043 at 2.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.18/0.22
Resolution: 2.40 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE MULTIPLE ANTIBIOTIC-RESISTANCE REPRESSOR (SSU05_1136) FROM STREPTOCOCCUS SUIS 89/1591 AT 2.20 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.22/0.28
Resolution: 2.20 Å
R/Rfree: 0.22/0.28
X-ray diffraction data for the Crystal structure of a protein of unknown function with a cystatin-like fold (npun_r3134) from nostoc punctiforme pcc 73102 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE AROMATIC RING HYDROXYLASE (SARO_3538) FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM AT 1.75 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.17/0.20
Resolution: 1.75 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of Pyridoxamine 5'-phosphate oxidase-like protein (NP_783940.1) from Lactobacillus plantarum at 1.55 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.55 Å
R/Rfree: 0.17/0.20
Resolution: 1.55 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a putative ketosteroid isomerase (sfri_1973) from shewanella frigidimarina ncimb 400 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.18/0.23
Resolution: 1.80 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the CRYSTAL STRUCTURE OF a DUF2118 family protein (MMP0046) FROM METHANOCOCCUS MARIPALUDIS AT 2.20 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.19/0.24
Resolution: 2.20 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of an ntf2-like protein with a cystatin-like fold (saro_3722) from novosphingobium aromaticivorans dsm at 1.16 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.16 Å
R/Rfree: 0.14/0.17
Resolution: 1.16 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE DEHYDRATASE FROM THE NTF2-LIKE FAMILY (SAV_4671) FROM STREPTOMYCES AVERMITILIS AT 2.10 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.20/0.24
Resolution: 2.10 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of putative acetyltransferase (NP_371943.1) from STAPHYLOCOCCUS AUREUS MU50 at 2.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.22/0.25
Resolution: 2.50 Å
R/Rfree: 0.22/0.25
X-ray diffraction data for the Crystal structure of a putative NTP pyrophosphohydrolase (Exig_1061) from EXIGUOBACTERIUM SP. 255-15 at 1.78 A resolution


First author:
G.W. Han
Resolution: 1.78 Å
R/Rfree: 0.18/0.22
Resolution: 1.78 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE SPORE COAT PROTEIN (BH2358) FROM BACILLUS HALODURANS C-125 AT 1.54 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.54 Å
R/Rfree: 0.16/0.19
Resolution: 1.54 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a putative AphA-like transcription factor (ZP_00208345.1) from Magnetospirillum magnetotacticum MS-1 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.21/0.26
Resolution: 2.00 Å
R/Rfree: 0.21/0.26
X-ray diffraction data for the Crystal structure of a putative kinase (caur_3907) from chloroflexus aurantiacus j-10-fl at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.18/0.22
Resolution: 1.70 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a putative 4-amino-4-deoxychorismate lyase (hs_0128) from haemophilus somnus 129pt at 2.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.19/0.23
Resolution: 2.40 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of Cysteine peptidase (NP_982244.1) from BACILLUS CEREUS ATCC 10987 at 2.50 A resolution


First author:
Q. Xu
Resolution: 2.50 Å
R/Rfree: 0.19/0.22
Resolution: 2.50 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of putative TetR transcriptional regulator (YP_510936.1) from Jannaschia sp. CCS1 at 1.79 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.79 Å
R/Rfree: 0.21/0.24
Resolution: 1.79 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of a methyltransferase-like protein (spo2022) from silicibacter pomeroyi dss-3 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.17/0.19
Resolution: 1.80 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of a domain of unknown function with a cystatin-like fold (npun_r1993) from nostoc punctiforme pcc 73102 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.16/0.22
Resolution: 2.00 Å
R/Rfree: 0.16/0.22
X-ray diffraction data for the Crystal structure of putative fructose transport system kinase (YP_612366.1) from Silicibacter sp. TM1040 at 1.95 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.17/0.22
Resolution: 1.95 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of a dj-1/pfpi-like protein (shew_2856) from shewanella loihica pv-4 at 1.76 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.76 Å
R/Rfree: 0.16/0.19
Resolution: 1.76 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a putative glutathione s-transferase (reut_a1011) from ralstonia eutropha jmp134 at 2.05 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.17/0.21
Resolution: 2.05 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of carboxylesterase (NP_108484.1) from Mesorhizobium loti at 1.75 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.16/0.19
Resolution: 1.75 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a putative f420 dependent nadp-reductase (arth_0613) from arthrobacter sp. fb24 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.16/0.20
Resolution: 1.70 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE ROSSMANN-LIKE DEHYDROGENASE (CGL2689) FROM CORYNEBACTERIUM GLUTAMICUM AT 2.07 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.07 Å
R/Rfree: 0.19/0.22
Resolution: 2.07 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a ring oxydation complex/ phenylacetic acid degradation-like protein (SSO1313) from Sulfolobus solfataricus P2 at 2.43 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.43 Å
R/Rfree: 0.18/0.20
Resolution: 2.43 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of a biotin protein ligase-like protein of unknown function (tm1040_0394) from silicibacter sp. tm1040 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.18/0.23
Resolution: 1.70 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of ribulose-5-phosphate 3-epimerase (YP_718263.1) from Haemophilus somnus 129PT at 1.91 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.91 Å
R/Rfree: 0.17/0.21
Resolution: 1.91 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a cadd-like protein of unknown function (npun_f6505) from nostoc punctiforme pcc 73102 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.17/0.23
Resolution: 2.00 Å
R/Rfree: 0.17/0.23
X-ray diffraction data for the CRYSTAL STRUCTURE OF A CADD-LIKE PROTEIN OF UNKNOWN FUNCTION (NPUN_F6505) FROM NOSTOC PUNCTIFORME PCC 73102 AT 1.35 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.35 Å
R/Rfree: 0.16/0.17
Resolution: 1.35 Å
R/Rfree: 0.16/0.17
X-ray diffraction data for the Crystal structure of a putative tetr/acrr family transcriptional regulator (saro_0558) from novosphingobium aromaticivorans dsm at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.18/0.21
Resolution: 1.80 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of predicted CIB-like hydrolase (NP_393672.1) from Thermoplasma acidophilum at 1.45 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.45 Å
R/Rfree: 0.16/0.19
Resolution: 1.45 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of IMP dehydrogenase/GMP reductase-like protein (NP_599840.1) from Corynebacterium glutamicum ATCC 13032 Kitasato at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE ALPHA-N-ACETYLGALACTOSAMINIDASE (BH1870) FROM BACILLUS HALODURANS C-125 AT 2.00 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.16/0.19
Resolution: 2.00 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE IRON-MOLYBDENUM COFACTOR (FEMO-CO) DINITROGENASE (TA1041M) FROM THERMOPLASMA ACIDOPHILUM DSM 1728 AT 1.30 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.16/0.18
Resolution: 1.30 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of putative MarR-like transcription regulator (NP_978771.1) from Bacillus cereus ATCC 10987 at 2.38 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.38 Å
R/Rfree: 0.23/0.25
Resolution: 2.38 Å
R/Rfree: 0.23/0.25
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE THIOESTERASE (TM1040_1390) FROM SILICIBACTER SP. TM1040 AT 2.15 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.20/0.25
Resolution: 2.15 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of a putativel NUDIX hydrolase (LMOf2365_2679) from Listeria monocytogenes str. 4b F2365 at 1.70 A resolution


First author:
Joint center for structural genomics (jcsg)
Resolution: 1.71 Å
R/Rfree: 0.16/0.20
Resolution: 1.71 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of a putative dinitrogenase (mj0327) from methanocaldococcus jannaschii dsm at 1.70 A resolution

First author:
K. Jaudzems
Resolution: 1.70 Å
R/Rfree: 0.20/0.26
Resolution: 1.70 Å
R/Rfree: 0.20/0.26
X-ray diffraction data for the Crystal structure of a putative nucleotidyltransferase (NP_343093.1) from Sulfolobus solfataricus at 1.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.15/0.19
Resolution: 1.40 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A putative acetyltransferase of the GNAT family (DDE_3044) FROM DESULFOVIBRIO DESULFURICANS SUBSP. AT 1.85 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.21/0.25
Resolution: 1.85 Å
R/Rfree: 0.21/0.25
X-ray diffraction data for the Crystal structure of a putative redox protein (lsei_0423) from lactobacillus casei atcc 334 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.14/0.16
Resolution: 1.50 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE DNA DAMAGE-INDUCIBLE PROTEIN (CHU_0679) FROM CYTOPHAGA HUTCHINSONII ATCC 33406 AT 1.50 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.16/0.19
Resolution: 1.50 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a putative acetyltransferase (DR_1678) from Deinococcus radiodurans R1 at 1.19 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.19 Å
R/Rfree: 0.14/0.15
Resolution: 1.19 Å
R/Rfree: 0.14/0.15
X-ray diffraction data for the Crystal structure of a putative yfit-like metal-dependent hydrolase (bh0186) from bacillus halodurans c-125 at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.15/0.20
Resolution: 2.30 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the Crystal structure of putative acetyltransferase (YP_831484.1) from Arthrobacter sp. FB24 at 1.65 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.65 Å
R/Rfree: 0.17/0.21
Resolution: 1.65 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of deoxyribonucleotidase-like protein (NP_764060.1) from Staphylococcus epidermidis ATCC 12228 at 1.55 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.55 Å
R/Rfree: 0.18/0.22
Resolution: 1.55 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a putative phosphoenolpyruvate phosphonomutase (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.14/0.17
Resolution: 1.80 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of PanE/ApbA family ketopantoate reductase (YP_299159.1) from Ralstonia eutropha JMP134 at 2.15 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.18/0.23
Resolution: 2.15 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of a putative fmn-dependent nitroreductase (ct0345) from chlorobium tepidum tls at 1.15 A resolution

First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.15 Å
R/Rfree: 0.15/0.17
Resolution: 1.15 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of a predicted dna-binding transcriptional regulator (saro_1072) from novosphingobium aromaticivorans dsm at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.17/0.20
Resolution: 2.10 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a predicted dna-binding transcriptional regulator (saro_1072) from novosphingobium aromaticivorans dsm at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.19/0.22
Resolution: 1.85 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE O-METHYLTRANSFERASE (NPUN_R0239) FROM NOSTOC PUNCTIFORME PCC 73102 AT 2.15 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.19/0.23
Resolution: 2.15 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE SUGAR PHOSPHATE ISOMERASE/EPIMERASE (AVA4194) FROM ANABAENA VARIABILIS ATCC 29413 AT 1.78 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.78 Å
R/Rfree: 0.18/0.22
Resolution: 1.78 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a putative hydrolase (ava_4197) from anabaena variabilis atcc 29413 at 1.35 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.35 Å
R/Rfree: 0.12/0.14
Resolution: 1.35 Å
R/Rfree: 0.12/0.14
X-ray diffraction data for the Crystal structure of a putative tena-like thiaminase (tena-1, sso2206) from sulfolobus solfataricus p2 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.17/0.18
Resolution: 1.50 Å
R/Rfree: 0.17/0.18
X-ray diffraction data for the Crystal structure of a putative lipase (NP_343859.1) from Sulfolobus solfataricus at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.16/0.19
Resolution: 1.85 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a two-domain protein containing dj-1/thij/pfpi-like and ferritin-like domains (ava_4496) from anabaena variabilis atcc 29413 at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.18/0.22
Resolution: 1.90 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of an uncharacterized protein (bh2621) from bacillus halodurans at 1.55 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.55 Å
R/Rfree: 0.18/0.21
Resolution: 1.55 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of putative dinucleotide-binding oxidoreductase (NP_786167.1) from Lactobacillus plantarum at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.15/0.19
Resolution: 1.60 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of putative metal-dependent hydrolase (YP_805737.1) from Lactobacillus casei ATCC 334 at 1.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.13/0.16
Resolution: 1.40 Å
R/Rfree: 0.13/0.16
X-ray diffraction data for the Crystal structure of a putative oxidoreductase (ZP_00056571.1) from Magnetospirillum magnetotacticum MS-1 at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.21/0.26
Resolution: 1.85 Å
R/Rfree: 0.21/0.26
X-ray diffraction data for the Crystal structure of protein of unknown function with ferritin-like fold (YP_832262.1) from Arthrobacter sp. FB24 at 2.33 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.33 Å
R/Rfree: 0.21/0.26
Resolution: 2.33 Å
R/Rfree: 0.21/0.26
X-ray diffraction data for the Crystal structure of a putative acetyltransferase (NP_394282.1) from Thermoplasma acidophilum at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.18/0.24
Resolution: 2.00 Å
R/Rfree: 0.18/0.24
X-ray diffraction data for the Crystal structure of a putative antidote protein of plasmid maintenance system (npun_f2943) from nostoc punctiforme pcc 73102 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.17/0.20
Resolution: 1.60 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the CRYSTAL STRUCTURE OF A RMLC-LIKE CUPIN (SFRI_3105) FROM SHEWANELLA FRIGIDIMARINA NCIMB 400 AT 1.90 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.19/0.26
Resolution: 1.90 Å
R/Rfree: 0.19/0.26
X-ray diffraction data for the Crystal structure of acetyltransferase GNAT family (NP_688560.1) from Streptococcus agalactiae 2603 at 1.28 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.28 Å
R/Rfree: 0.17/0.18
Resolution: 1.28 Å
R/Rfree: 0.17/0.18
X-ray diffraction data for the Crystal structure of Thioesterase superfamily protein (ZP_00837258.1) from Shewanella loihica PV-4 at 1.65 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.20/0.28
Resolution: 1.50 Å
R/Rfree: 0.20/0.28
X-ray diffraction data for the Crystal structure of putative thioesterase (YP_496845.1) from Novosphingobium aromaticivorans DSM 12444 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.17/0.20
Resolution: 1.70 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE THIOESTERASE II (TFU_2367) FROM THERMOBIFIDA FUSCA YX AT 2.45 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.16 Å
R/Rfree: 0.18/0.24
Resolution: 2.16 Å
R/Rfree: 0.18/0.24
X-ray diffraction data for the Crystal structure of a putative endoribonuclease (so_1960) from shewanella oneidensis mr-1 at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.15/0.18
Resolution: 1.85 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of acetyltransferase GNAT family (YP_013287.1) from Listeria monocytogenes 4b F2365 at 1.46 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.46 Å
R/Rfree: 0.17/0.19
Resolution: 1.46 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of a histidine triad (hit) protein (mfla_2506) from methylobacillus flagellatus kt at 1.65 A resolution

First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.65 Å
R/Rfree: 0.18/0.22
Resolution: 1.65 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of an osmc-like protein (gsu2788) from geobacter sulfurreducens at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.15/0.17
Resolution: 1.50 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of a putative thioesterase (tm1040_2492) from silicibacter sp. tm1040 at 1.79 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.79 Å
R/Rfree: 0.22/0.27
Resolution: 1.79 Å
R/Rfree: 0.22/0.27
X-ray diffraction data for the Crystal structure of a xylose isomerase domain containing protein (stm4435) from salmonella typhimurium lt2 at 2.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.18/0.23
Resolution: 2.40 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of an osmc-like hydroperoxide resistance protein (jann_2040) from jannaschia sp. ccs1 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.14/0.16
Resolution: 1.70 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of a Queuosine biosynthesis protein queC (ECA1155) from Erwinia carotovora subsp. atroseptica SCRI1043 at 2.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.19/0.22
Resolution: 2.40 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE MANNOSE-6-PHOSPHATE ISOMERASE (REUT_A1446) FROM RALSTONIA EUTROPHA JMP134 AT 2.10 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of GCN5-related N-acetyltransferase (YP_295895.1) from Ralstonia eutropha JMP134 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.14/0.17
Resolution: 1.80 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of multiple antibiotic-resistance repressor (MarR) (YP_013417.1) from Listeria monocytogenes 4b F2365 at 2.07 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.07 Å
R/Rfree: 0.24/0.29
Resolution: 2.07 Å
R/Rfree: 0.24/0.29
X-ray diffraction data for the Crystal structure of a putative acetyltransferase belonging to the gnat family (xcc2953) from xanthomonas campestris pv. campestris at 1.40 A resolution

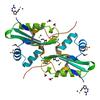
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.14/0.16
Resolution: 1.40 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of a cupin-2 domain containing protein (sfri_3543) from shewanella frigidimarina ncimb 400 at 2.05 A resolution

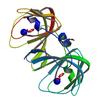
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.19/0.21
Resolution: 2.05 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the Crystal structure of an rmlc-like cupin protein (reut_a0381) from ralstonia eutropha jmp134 at 2.60 A resolution

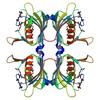
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.60 Å
R/Rfree: 0.23/0.26
Resolution: 2.60 Å
R/Rfree: 0.23/0.26
X-ray diffraction data for the Crystal structure of haloacid delahogenase-like family hydrolase (NP_639141.1) from Xanthomonas campestris at 1.81 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.81 Å
R/Rfree: 0.18/0.23
Resolution: 1.81 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of Uncharacterized peroxidase-related protein (YP_604910.1) from Deinococcus geothermalis DSM 11300 at 1.51 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.51 Å
R/Rfree: 0.18/0.22
Resolution: 1.51 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of an ADP-ribose pyrophosphatase (SSU98_1448) from STREPTOCOCCUS SUIS 89-1591 at 2.27 A resolution

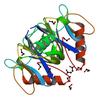
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.27 Å
R/Rfree: 0.19/0.22
Resolution: 2.27 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF a SAM dependent methyl-transferase type 12 family protein (ECA1738) FROM PECTOBACTERIUM ATROSEPTICUM SCRI1043 AT 1.74 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.74 Å
R/Rfree: 0.16/0.18
Resolution: 1.74 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of a sam dependent methyl-transferase type 12 family protein (eca1738) from pectobacterium atrosepticum scri1043 at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.15/0.18
Resolution: 1.85 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE THIOESTERASE, PHENYLACETIC ACID DEGRADATION-RELATED PROTEIN (REUT_B4779) FROM RALSTONIA EUTROPHA JMP134 AT 2.20 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.18/0.22
Resolution: 2.20 Å
R/Rfree: 0.18/0.22