6726 results
X-ray diffraction data for the Motor domain of human kinesin family member C1


First author:
H.W. Park
Resolution: 2.25 Å
R/Rfree: 0.22/0.27
Resolution: 2.25 Å
R/Rfree: 0.22/0.27
X-ray diffraction data for the Crystal structure of hybrid tudor domain of human lysine demethylase KDM4B


First author:
F. Wang
Resolution: 2.56 Å
R/Rfree: 0.20/0.25
Resolution: 2.56 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of double tudor domain of human lysine demethylase KDM4A complexed with histone H3K23me3


First author:
F. Wang
Resolution: 2.29 Å
R/Rfree: 0.26/0.30
Resolution: 2.29 Å
R/Rfree: 0.26/0.30
X-ray diffraction data for the Crystal structure of double tudor domain of human lysine demethylase KDM4A


First author:
F. Wang
Resolution: 2.15 Å
R/Rfree: 0.16/0.20
Resolution: 2.15 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of double tudor domain of human lysine demethylase KDM4A


First author:
F. Wang
Resolution: 1.99 Å
R/Rfree: 0.17/0.20
Resolution: 1.99 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of an N-terminal HTATIP fragment

First author:
Y. Liu
Resolution: 2.80 Å
R/Rfree: 0.23/0.26
Resolution: 2.80 Å
R/Rfree: 0.23/0.26
X-ray diffraction data for the Crystal structure of hemagglutinin from H1N1 Influenza A virus A/Denver/57 bound to the C05 antibody


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.92 Å
R/Rfree: 0.20/0.26
Resolution: 2.92 Å
R/Rfree: 0.20/0.26
X-ray diffraction data for the X-ray Crystal Structure of a Putative Phosphate ABC Transporter Substrate-Binding Protein with Bound Phosphate from Clostridium perfringens


First author:
J.S. Brunzelle
Gene name: pstS
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
Gene name: pstS
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the WWE domain of human HUWE1

First author:
L. Halabelian
Resolution: 2.00 Å
R/Rfree: 0.18/0.21
Resolution: 2.00 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of putative Xaa-His dipeptidase (YP_718209.1) from Haemophilus somnus 129PT at 2.11 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.11 Å
R/Rfree: 0.22/0.24
Resolution: 2.11 Å
R/Rfree: 0.22/0.24
X-ray diffraction data for the Crystal structure of a succinyl-diaminopimelate desuccinylase (ArgE) from Corynebacterium glutamicum ATCC 13032 at 2.97 A resolution


First author:
A.T. Brunger
Resolution: 2.97 Å
R/Rfree: 0.24/0.26
Resolution: 2.97 Å
R/Rfree: 0.24/0.26
X-ray diffraction data for the Crystal structure of a tripeptidase (SAV1512) from staphylococcus aureus subsp. aureus mu50 at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of aminopeptidase PepT (NP_980509.1) from Bacillus cereus ATCC 10987 at 2.04 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.04 Å
R/Rfree: 0.17/0.21
Resolution: 2.04 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of co-catalytic metallopeptidase (YP_387682.1) from Desulfovibrio desulfuricans G20 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the CRYSTAL STRUCTURE OF a putative metallopeptidase (SDEN_2526) FROM SHEWANELLA DENITRIFICANS OS217 AT 1.74 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.74 Å
R/Rfree: 0.18/0.22
Resolution: 1.74 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of Putative metallopeptidase (YP_001051774.1) from SHEWANELLA BALTICA OS155 at 2.45 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.45 Å
R/Rfree: 0.16/0.22
Resolution: 2.45 Å
R/Rfree: 0.16/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE METALLOPEPTIDASE (SAMA_0725) FROM SHEWANELLA AMAZONENSIS SB2B AT 2.00 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.16/0.21
Resolution: 2.00 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of putative M42 glutamyl aminopeptidase (YP_676701.1) from Cytophaga hutchinsonii ATCC 33406 at 2.39 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.39 Å
R/Rfree: 0.19/0.22
Resolution: 2.39 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a leucine aminopeptidase precursor (BT_2548) from Bacteroides thetaiotaomicron VPI-5482 at 1.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.12/0.16
Resolution: 1.30 Å
R/Rfree: 0.12/0.16
X-ray diffraction data for the Crystal structure of Putative succinylglutamate desuccinylase/aspartoacylase (YP_749235.1) from Shewanella frigidimarinA NCIMB 400 at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.16/0.19
Resolution: 2.10 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF a putative succinylglutamate desuccinylase / aspartoacylase family protein (SAMA_0604) FROM SHEWANELLA AMAZONENSIS SB2B AT 1.80 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.16/0.19
Resolution: 1.80 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of Putative carboxypeptidase A (YP_562911.1) from SHEWANELLA DENITRIFICANS OS-217 at 2.39 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.39 Å
R/Rfree: 0.18/0.23
Resolution: 2.39 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of putative carboxypeptidase (YP_103406.1) from BURKHOLDERIA MALLEI ATCC 23344 at 2.49 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.49 Å
R/Rfree: 0.16/0.19
Resolution: 2.49 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a putative zinc peptidase (NP_812461.1) from Bacteroides thetaiotaomicron VPI-5482 at 2.31 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.31 Å
R/Rfree: 0.19/0.25
Resolution: 2.31 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the Crystal structure of putative peptidase (NP_348812.1) from CLOSTRIDIUM ACETOBUTYLICUM at 2.37 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.37 Å
R/Rfree: 0.17/0.21
Resolution: 2.37 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a succinylglutamate desuccinylase (TM1040_2694) from SILICIBACTER SP. TM1040 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.16/0.20
Resolution: 2.00 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of an aspartoacylase family protein (mlr6093) from mesorhizobium loti maff303099 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.21/0.25
Resolution: 2.00 Å
R/Rfree: 0.21/0.25
X-ray diffraction data for the Crystal Structure of the SRAP Domain of Human HMCES Protein


First author:
L. Halabelian
Resolution: 1.50 Å
R/Rfree: 0.18/0.21
Resolution: 1.50 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the The tetramer structure of the nervy homolgy two (NHR2) domain of AML1-ETO is critical for AML1-ETO'S activity


First author:
Y. Liu
Resolution: 2.00 Å
R/Rfree: 0.21/0.27
Resolution: 2.00 Å
R/Rfree: 0.21/0.27
X-ray diffraction data for the Crystal structure of carbon-nitrogen hydrolase from Helicobacter pylori G27


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a low occupancy fragment candidate (5-[(4-Isopropylphenyl)amino]-6-methyl-1,2,4-triazin-3(2H)-one) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain

First author:
R.J. Harding
Resolution: 1.07 Å
R/Rfree: 0.14/0.15
Resolution: 1.07 Å
R/Rfree: 0.14/0.15
X-ray diffraction data for the Crystal structure of a low occupancy fragment candidate (N-(4-Methyl-1,3-thiazol-2-yl)propanamide) bound adjacent to the ubiquitin binding pocket of the HDAC6 zinc-finger domain

First author:
R.J. Harding
Resolution: 1.05 Å
R/Rfree: 0.13/0.15
Resolution: 1.05 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Crystal structure of fragment (3-[6-Oxo-3-(3-pyridinyl)-1(6H)-pyridazinyl]propanoic acid) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain

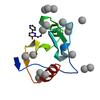
First author:
R.J. Harding
Resolution: 1.70 Å
R/Rfree: 0.15/0.18
Resolution: 1.70 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of fragment (3-(5-Chloro-1,3-benzothiazol-2-yl)propanoic acid) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain

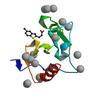
First author:
R.J. Harding
Resolution: 1.60 Å
R/Rfree: 0.16/0.19
Resolution: 1.60 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of fragment 3-(1,3-Benzothiazol-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain

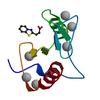
First author:
R.J. Harding
Resolution: 1.80 Å
R/Rfree: 0.15/0.18
Resolution: 1.80 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of fragment 3-(1-Methyl-2-oxo-1,2-dihydroquinoxalin-3-yl)propionic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain

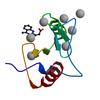
First author:
R.J. Harding
Resolution: 1.55 Å
R/Rfree: 0.15/0.17
Resolution: 1.55 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of fragment 3-(3-Methyl-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain

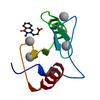
First author:
R.J. Harding
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of fragment 3-(3-Methoxy-2-quinoxalinyl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain

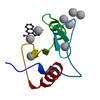
First author:
R.J. Harding
Resolution: 1.55 Å
R/Rfree: 0.16/0.19
Resolution: 1.55 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of fragment 3-(quinolin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain

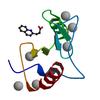
First author:
R.J. Harding
Resolution: 1.60 Å
R/Rfree: 0.16/0.20
Resolution: 1.60 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of fragment 2-(Benzo[d]thiazol-2-yl)acetic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain

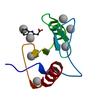
First author:
R.J. Harding
Resolution: 1.55 Å
R/Rfree: 0.15/0.17
Resolution: 1.55 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Structure of HDAC6 zinc-finger ubiquitin binding domain soaked with 3,3'-(benzo[1,2-d:5,4-d']bis(thiazole)-2,6-diyl)dipropionic acid


First author:
R.J. Harding
Resolution: 1.60 Å
R/Rfree: 0.17/0.20
Resolution: 1.60 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE PROTEASE (BDI_1141) FROM PARABACTEROIDES DISTASONIS ATCC 8503 AT 2.00 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.17/0.20
Resolution: 2.00 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a susd homolog (bt_3984) from bacteroides thetaiotaomicron vpi-5482 at 1.70 A resolution


First author:
C. Bakolitsa
Resolution: 1.70 Å
R/Rfree: 0.14/0.17
Resolution: 1.70 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of a putative outer membrane protein, probably involved in nutrient binding (BVU_1254) from Bacteroides vulgatus ATCC 8482 at 1.42 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.42 Å
R/Rfree: 0.14/0.17
Resolution: 1.42 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of a susd homolog (BVU_2203) from Bacteroides vulgatus ATCC 8482 at 1.85 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.14/0.16
Resolution: 1.85 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of a SusD-like protein (BT_3752) from Bacteroides thetaiotaomicron VPI-5482 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.14/0.17
Resolution: 1.70 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the CRYSTAL STRUCTURE OF A SUSD HOMOLOG (BF3025) FROM BACTEROIDES FRAGILIS NCTC 9343 AT 1.50 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.15/0.17
Resolution: 1.50 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of a SusD homolog (BF0972) from Bacteroides fragilis NCTC 9343 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.16/0.22
Resolution: 2.00 Å
R/Rfree: 0.16/0.22
X-ray diffraction data for the Crystal structure of a susd superfamily protein (bf3413) from bacteroides fragilis nctc 9343 at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.09 Å
R/Rfree: 0.16/0.20
Resolution: 2.09 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of a putative sel1 repeat protein (kpn_04481) from Klebsiella pneumoniae subsp. pneumoniae at 1.65 a resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.65 Å
R/Rfree: 0.17/0.19
Resolution: 1.65 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of a putative cell wall hydrolase (CD630_03720) from Clostridium difficile 630 at 2.38 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.38 Å
R/Rfree: 0.18/0.21
Resolution: 2.38 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a putative calcium-regulated periplasmic protein (bt0923) from bacteroides thetaiotaomicron at 1.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.16/0.20
Resolution: 1.40 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of an Uncharacterized DUF3829-like protein (BT_1908) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.21/0.24
Resolution: 2.10 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of a putative membrane protein of unknown function (yfdx) from klebsiella pneumoniae subsp. at 1.65 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.65 Å
R/Rfree: 0.15/0.19
Resolution: 1.65 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a lipoprotein YedD (KPN_02420) from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 at 1.38 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.38 Å
R/Rfree: 0.15/0.18
Resolution: 1.38 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative hydrolase (BT_2193) from Bacteroides thetaiotaomicron VPI-5482 at 1.25 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.25 Å
R/Rfree: 0.15/0.18
Resolution: 1.25 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of glycosyl hydrolase family 5 (NP_809925.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.19/0.22
Resolution: 2.10 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a fimbrial assembly protein (BDI_3522) from Parabacteroides distasonis ATCC 8503 at 2.38 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.38 Å
R/Rfree: 0.23/0.25
Resolution: 2.38 Å
R/Rfree: 0.23/0.25
X-ray diffraction data for the Crystal structure of a putative cell adhesion protein (BT0320) from Bacteroides thetaiotaomicron VPI-5482 at 2.37 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.37 Å
R/Rfree: 0.19/0.22
Resolution: 2.37 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of an outer membrane protein, part of a putative carbohydrate binding complex (bt_1022) from bacteroides thetaiotaomicron vpi-5482 at 1.93 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.93 Å
R/Rfree: 0.20/0.22
Resolution: 1.93 Å
R/Rfree: 0.20/0.22
X-ray diffraction data for the Crystal structure of Putative sugar binding protein (YP_001300177.1) from Bacteroides vulgatus ATCC 8482 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a putative cell adhesion protein (BF2867) from Bacteroides fragilis NCTC 9343 at 2.57 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.57 Å
R/Rfree: 0.19/0.23
Resolution: 2.57 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of a member of duf3298 family (BF2082) from bacteroides fragilis nctc 9343 at 2.30 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.19/0.21
Resolution: 2.30 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the CRYSTAL STRUCTURE OF AN ABC TRANSPORTER (BDI_1369) FROM PARABACTEROIDES DISTASONIS AT 1.75 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.17/0.21
Resolution: 1.75 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of putative glucoamylase (YP_210071.1) from Bacteroides fragilis NCTC 9343 at 2.12 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.12 Å
R/Rfree: 0.16/0.21
Resolution: 2.12 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of a putative nucleoside deoxyribosyltransferase (BDI_0649) from Parabacteroides distasonis ATCC 8503 at 2.40 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.19/0.21
Resolution: 2.40 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the Crystal structure of a fimbrial protein (BVU_2522) from Bacteroides vulgatus ATCC 8482 at 2.55 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.55 Å
R/Rfree: 0.17/0.20
Resolution: 2.55 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of Putative Peptide:N-glycosidase F (PNGase F) (YP_210507.1) from Bacteroides fragilis NCTC 9343 at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.20/0.24
Resolution: 2.30 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of a putative hydrolase (BVU_2763) from Bacteroides vulgatus ATCC 8482 at 2.44 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.44 Å
R/Rfree: 0.17/0.22
Resolution: 2.44 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of periplasmic sugar-binding protein (YP_001338366.1) from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 at 1.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.16/0.17
Resolution: 1.30 Å
R/Rfree: 0.16/0.17
X-ray diffraction data for the Crystal structure of Putative lipid binding protein (YP_001304415.1) from Parabacteroides distasonis ATCC 8503 at 2.16 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.16 Å
R/Rfree: 0.19/0.21
Resolution: 2.16 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the Crystal structure of a glycoside hydrolase family 5 (BVU_2644) from Bacteroides vulgatus ATCC 8482 at 1.90 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.14/0.17
Resolution: 1.90 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of a putative glycosyl hydrolase (BT3469) from Bacteroides thetaiotaomicron VPI-5482 at 2.49 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.49 Å
R/Rfree: 0.17/0.21
Resolution: 2.49 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of putative alpha-L-fucosidase (NP_812709.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.14/0.17
Resolution: 1.60 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE LIPID BINDING PROTEIN (BT_2261) FROM BACTEROIDES THETAIOTAOMICRON VPI-5482 AT 2.20 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.19/0.23
Resolution: 2.20 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of putative polysaccharide binding proteins (DUF1812) (NP_809975.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.20 A resolution


First author:
Q. Xu
Resolution: 2.20 Å
R/Rfree: 0.19/0.23
Resolution: 2.20 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of Putative glycosyl hydrolase (YP_001301887.1) from Parabacteroides distasonis ATCC 8503 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.16/0.22
Resolution: 2.00 Å
R/Rfree: 0.16/0.22
X-ray diffraction data for the Crystal structure of a putative dna binding protein (bt_1116) from bacteroides thetaiotaomicron vpi-5482 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.14/0.17
Resolution: 1.50 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of Putative cell adhesion protein (YP_001304840.1) from Parabacteroides distasonis ATCC 8503 at 2.05 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.17/0.22
Resolution: 2.05 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of a Putative lipoprotein (BF3042) from Bacteroides fragilis NCTC 9343 at 1.87 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.87 Å
R/Rfree: 0.19/0.22
Resolution: 1.87 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a putative lipoprotein (BDI_3050) from Parabacteroides distasonis ATCC 8503 at 2.70 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.70 Å
R/Rfree: 0.19/0.22
Resolution: 2.70 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE EXPORTED PROTEIN WITH YMCC-LIKE FOLD (BF2203) FROM BACTEROIDES FRAGILIS NCTC 9343 AT 2.22 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.22 Å
R/Rfree: 0.20/0.25
Resolution: 2.22 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of putative beta-lactamase (NP_815223.1) from Enterococcus faecalis V583 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.15/0.17
Resolution: 1.50 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of a putative sugar binding protein (BT_4411) from Bacteroides thetaiotaomicron VPI-5482 at 1.25 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.25 Å
R/Rfree: 0.13/0.15
Resolution: 1.25 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Crystal structure of putative membrane-associated protein of unknown function (YP_211325.1) from Bacteroides fragilis NCTC 9343 at 2.20 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.17/0.21
Resolution: 2.20 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of Putative pore-forming toxin (YP_001301288.1) from Bacteroides vulgatus ATCC 8482 at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.18/0.21
Resolution: 1.85 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of putative adhesin (YP_001304413.1) from Parabacteroides distasonis ATCC 8503 at 2.41 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.41 Å
R/Rfree: 0.19/0.23
Resolution: 2.41 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of putative nucleic acid-binding lipoprotein (YP_001337197.1) from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 at 2.46 A resolution


First author:
D. Das
Resolution: 2.46 Å
R/Rfree: 0.19/0.23
Resolution: 2.46 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of Putative pectinase (YP_001304412.1) from Parabacteroides distasonis ATCC 8503 at 2.30 A resolution

First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.24/0.28
Resolution: 2.30 Å
R/Rfree: 0.24/0.28
X-ray diffraction data for the Crystal structure of a putative member of duf3244 protein family (BT_3571) from Bacteroides thetaiotaomicron vpi-5482 at 1.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.16/0.20
Resolution: 1.40 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of protein of unknown function DUF1344 (YP_001299214.1) from Bacteroides vulgatus ATCC 8482 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.18/0.21
Resolution: 2.00 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a putative acetyl xylan esterase (BF1801) from Bacteroides fragilis NCTC 9343 at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.15/0.18
Resolution: 2.30 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE HYDROLASE (BVU_4111) FROM BACTEROIDES VULGATUS ATCC 8482 AT 1.90 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.17/0.21
Resolution: 1.90 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a porin-like protein (BVU_2266) from Bacteroides vulgatus ATCC 8482 at 2.80 A resolution


First author:
Joint center for structural genomics (JCSG)
Resolution: 2.80 Å
R/Rfree: 0.20/0.24
Resolution: 2.80 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of a Putative bacterial DNA binding protein (BVU_2165) from Bacteroides vulgatus ATCC 8482 at 2.25 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.25 Å
R/Rfree: 0.17/0.21
Resolution: 2.25 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a SusD homolog (BDI_0600) from Parabacteroides distasonis ATCC 8503 AT 2.05 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.24/0.27
Resolution: 2.05 Å
R/Rfree: 0.24/0.27
X-ray diffraction data for the Crystal structure of a putative cell adhesion protein (BDI_3519) from Parabacteroides distasonis ATCC 8503 at 2.34 A resolution (PSI Community Target, Nakayama)


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.34 Å
R/Rfree: 0.18/0.23
Resolution: 2.34 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of a putative glycosyl hydrolase (BDI_2473) from Parabacteroides distasonis ATCC 8503 at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.17/0.20
Resolution: 1.85 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a putative glycosyl hydrolase (BDI_3914) from Parabacteroides distasonis ATCC 8503 at 2.13 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.13 Å
R/Rfree: 0.17/0.20
Resolution: 2.13 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a PDZ domain protein (BDI_1242) from Parabacteroides distasonis ATCC 8503 at 2.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.19/0.22
Resolution: 2.50 Å
R/Rfree: 0.19/0.22