1160 results
X-ray diffraction data for the Crystal Structure of Hexapeptide-Repeat containing-Acetyltransferase VCA0836 Complexed with Acetyl Co Enzyme A from Vibrio cholerae O1 biovar eltor

First author:
Y. Kim
Resolution: 2.35 Å
R/Rfree: 0.19/0.25
Resolution: 2.35 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the X-ray crystal structure of Listeria monocytogenes EGD-e UDP-N-acetylenolpyruvylglucosamine reductase (MurB)

First author:
E.V. Filippova
Gene name: -
Resolution: 2.69 Å
R/Rfree: 0.21/0.27
Gene name: -
Resolution: 2.69 Å
R/Rfree: 0.21/0.27
X-ray diffraction data for the Crystal structure of an alpha subunit of tryptophan synthase from Vibrio cholerae O1 biovar El Tor str. N16961

First author:
B. Nocek
Resolution: 2.10 Å
R/Rfree: 0.18/0.23
Resolution: 2.10 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal Structure of Uridine Phosphorylase Complexed with Uracil from Vibrio cholerae O1 biovar El Tor

First author:
N. Maltseva
Resolution: 2.00 Å
R/Rfree: 0.17/0.23
Resolution: 2.00 Å
R/Rfree: 0.17/0.23
X-ray diffraction data for the 2.75 Angstrom Crystal Structure of Hypothetical Protein lmo2686 from Listeria monocytogenes EGD-e

First author:
G. Minasov
Gene name: -
Resolution: 2.75 Å
R/Rfree: 0.20/0.24
Gene name: -
Resolution: 2.75 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the 1.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus (IDP00699) in complex with NAD+

First author:
G.Minasov A.S.Halavaty
Gene name: betB
Resolution: 1.85 Å
R/Rfree: 0.13/0.17
Gene name: betB
Resolution: 1.85 Å
R/Rfree: 0.13/0.17
X-ray diffraction data for the NH3-dependent NAD synthetase from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with the nitrate ion

First author:
E.V. Filippova
Gene name: nadE
Resolution: 2.74 Å
R/Rfree: 0.21/0.24
Gene name: nadE
Resolution: 2.74 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of glutamate racemase from Listeria monocytogenes

First author:
K.A. Majorek
Gene name: racE
Resolution: 2.30 Å
R/Rfree: 0.19/0.24
Gene name: racE
Resolution: 2.30 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii

First author:
S.M. Anderson
Gene name: tpiA
Resolution: 2.50 Å
R/Rfree: 0.17/0.21
Gene name: tpiA
Resolution: 2.50 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Structure of the Salmonella enterica N-acetylmannosamine-6-phosphate 2-epimerase

First author:
S.M. Anderson
Gene name: nanE
Resolution: 1.50 Å
R/Rfree: 0.15/0.18
Gene name: nanE
Resolution: 1.50 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the 1.1 Angstrom Crystal Structure of Hypothetical Protein BA_2335 from Bacillus anthracis

First author:
G. Minasov
Resolution: 1.10 Å
R/Rfree: 0.15/0.18
Resolution: 1.10 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the 1.8 Angstrom Resolution Crystal Structure of Enoyl-CoA Hydratase from Bacillus anthracis.

First author:
G. Minasov
Resolution: 1.80 Å
R/Rfree: 0.16/0.19
Resolution: 1.80 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of glutamate racemase from Listeria monocytogenes in complex with acetate ion

First author:
K.A. Majorek
Gene name: racE
Resolution: 1.85 Å
R/Rfree: 0.17/0.22
Gene name: racE
Resolution: 1.85 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of the 3-oxoacyl-acyl carrier protein reductase, FabG, from Staphylococcus aureus

First author:
S.M. Anderson
Gene name: fabG
Resolution: 1.90 Å
R/Rfree: 0.16/0.19
Gene name: fabG
Resolution: 1.90 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the 2.00 Angstrom resolution crystal structure of a quinolinate phosphoribosyltransferase from Vibrio cholerae O1 biovar eltor str. N16961

First author:
A.S. Halavaty
Resolution: 2.00 Å
R/Rfree: 0.17/0.22
Resolution: 2.00 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Structure of 3-ketoacyl-(acyl-carrier-protein)reductase (FabG) from Vibrio cholerae O1 complexed with NADP+ (space group P62)

First author:
J. Hou
Gene name: fabG
Resolution: 1.95 Å
R/Rfree: 0.13/0.16
Gene name: fabG
Resolution: 1.95 Å
R/Rfree: 0.13/0.16
X-ray diffraction data for the Structure of the YghA Oxidoreductase from Salmonella enterica

First author:
S.M. Anderson
Gene name: yghA
Resolution: 1.25 Å
R/Rfree: 0.13/0.15
Gene name: yghA
Resolution: 1.25 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Structural flexibility in region involved in dimer formation of nuclease domain of Ribonuclase III (rnc) from Campylobacter jejuni

First author:
G. Minasov
Gene name: rnc
Resolution: 1.25 Å
R/Rfree: 0.13/0.17
Gene name: rnc
Resolution: 1.25 Å
R/Rfree: 0.13/0.17
X-ray diffraction data for the 2.4 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii.

First author:
G. Minasov
Resolution: 2.40 Å
R/Rfree: 0.19/0.24
Resolution: 2.40 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of 4-hydroxythreonine-4-phosphate dehydrogenase from Yersinia pestis CO92

First author:
B. Nocek
Gene name: pdxA
Resolution: 1.70 Å
R/Rfree: 0.19/0.25
Gene name: pdxA
Resolution: 1.70 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, complexed with IMP.

First author:
N.Maltseva 'J.Osipiuk
Gene name: guaB
Resolution: 2.15 Å
R/Rfree: 0.21/0.26
Gene name: guaB
Resolution: 2.15 Å
R/Rfree: 0.21/0.26
X-ray diffraction data for the 2.65 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose reductase (rfbD) from Bacillus anthracis str. Ames in complex with NADP

First author:
A.S. Halavaty
Gene name: rfbD
Resolution: 2.65 Å
R/Rfree: 0.22/0.26
Gene name: rfbD
Resolution: 2.65 Å
R/Rfree: 0.22/0.26
X-ray diffraction data for the 2.1 Angstrom resolution crystal structure of glycerol-3-phosphate dehydrogenase (gpsA) from Coxiella burnetii

First author:
G. Minasov
Gene name: gpsA
Resolution: 2.10 Å
R/Rfree: 0.16/0.21
Gene name: gpsA
Resolution: 2.10 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Structure of 3-oxoacyl-acylcarrier protein reductase, FabG from Francisella tularensis

First author:
S.M. Anderson
Gene name: fabG
Resolution: 1.95 Å
R/Rfree: 0.20/0.24
Gene name: fabG
Resolution: 1.95 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the 1.5A Crystal Structure of a Putative Peptidase E Protein from Listeria monocytogenes EGD-e

First author:
J.S. Brunzelle
Resolution: 1.50 Å
R/Rfree: 0.17/0.22
Resolution: 1.50 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis.

First author:
G. Minasov
Resolution: 2.20 Å
R/Rfree: 0.18/0.21
Resolution: 2.20 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the 1.54A Resolution Crystal Structure of a Beta-Carbonic Anhydrase from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2

First author:
J.S. Brunzelle
Gene name: yadF
Resolution: 1.54 Å
R/Rfree: 0.16/0.17
Gene name: yadF
Resolution: 1.54 Å
R/Rfree: 0.16/0.17
X-ray diffraction data for the 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Salmonella typhimurium

First author:
J. Osipiuk
Gene name: ispF
Resolution: 2.03 Å
R/Rfree: 0.17/0.21
Gene name: ispF
Resolution: 2.03 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of divalent-cation tolerance protein CutA from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2

First author:
B. Nocek
Gene name: cutA
Resolution: 1.90 Å
R/Rfree: 0.18/0.22
Gene name: cutA
Resolution: 1.90 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of bifunctional folylpolyglutamate synthase/dihydrofolate synthase with Mn, AMPPNP and L-Glutamate bound

First author:
B. Nocek
Gene name: folC
Resolution: 2.00 Å
R/Rfree: 0.17/0.20
Gene name: folC
Resolution: 2.00 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of Acetoacetyl-CoA thiolase (thlA2) from Clostridium difficile

First author:
S.M. Anderson
Gene name: thlA2
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
Gene name: thlA2
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the 2.0 Angstrom Resolution Crystal Structure of Glucose-6-phosphate Isomerase (pgi) from Bacillus anthracis.

First author:
G. Minasov
Gene name: pgi
Resolution: 2.00 Å
R/Rfree: 0.14/0.19
Gene name: pgi
Resolution: 2.00 Å
R/Rfree: 0.14/0.19
X-ray diffraction data for the Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure.

First author:
E.V. Filippova
Gene name: purE
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
Gene name: purE
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the 2.6 Angstrom Crystal Structure of Putative yceG-like Protein lmo1499 from Listeria monocytogenes

First author:
G. Minasov
Gene name: -
Resolution: 2.60 Å
R/Rfree: 0.17/0.22
Gene name: -
Resolution: 2.60 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL

First author:
A.S. Halavaty
Gene name: alr
Resolution: 2.37 Å
R/Rfree: 0.20/0.26
Gene name: alr
Resolution: 2.37 Å
R/Rfree: 0.20/0.26
X-ray diffraction data for the 2.1 Angstrom resolution crystal structure of an Orotate Phosphoribosyltransferase (pyrE) from Vibrio cholerae O1 biovar eltor str. N16961

First author:
A.S. Halavaty
Gene name: pyrE
Resolution: 2.10 Å
R/Rfree: 0.22/0.27
Gene name: pyrE
Resolution: 2.10 Å
R/Rfree: 0.22/0.27
X-ray diffraction data for the 1.63 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose 3,5-epimerase (rfbC) from Bacillus anthracis str. Ames with TDP and PPi bound

First author:
A.S. Halavaty
Gene name: rfbC
Resolution: 1.63 Å
R/Rfree: 0.12/0.16
Gene name: rfbC
Resolution: 1.63 Å
R/Rfree: 0.12/0.16
X-ray diffraction data for the Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant

First author:
J. Osipiuk
Gene name: guaB
Resolution: 2.16 Å
R/Rfree: 0.17/0.19
Gene name: guaB
Resolution: 2.16 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the 1.45 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with Shikimate and NAD.

First author:
G. Minasov
Gene name: -
Resolution: 1.45 Å
R/Rfree: 0.15/0.18
Gene name: -
Resolution: 1.45 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the UDP-N-acetylglucosamine acyltransferase from Campylobacter jejuni

First author:
J. Osipiuk
Gene name: lpxA
Resolution: 2.30 Å
R/Rfree: 0.20/0.24
Gene name: lpxA
Resolution: 2.30 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the D-Erythronate-4-Phosphate Dehydrogenase complexed with NAD

First author:
E.V. Filippova
Gene name: pdxB
Resolution: 2.36 Å
R/Rfree: 0.19/0.25
Gene name: pdxB
Resolution: 2.36 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with AMP and Succinate

First author:
N. Maltseva
Gene name: purB
Resolution: 1.92 Å
R/Rfree: 0.17/0.20
Gene name: purB
Resolution: 1.92 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis

First author:
N. Maltseva
Gene name: panC
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
Gene name: panC
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of putative 3-ketoacyl-(acyl-carrier-protein) reductase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with NADP+

First author:
J. Hou
Gene name: fabG
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
Gene name: fabG
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of transcriptional regulator VanUg, Form II

First author:
P.J. Stogios
Gene name: vanUG
Resolution: 1.12 Å
R/Rfree: 0.14/0.16
Gene name: vanUG
Resolution: 1.12 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of the Salmonella typhimurium FadA 3-ketoacyl-CoA thiolase

First author:
S.M. Anderson
Gene name: fadA
Resolution: 1.70 Å
R/Rfree: 0.14/0.19
Gene name: fadA
Resolution: 1.70 Å
R/Rfree: 0.14/0.19
X-ray diffraction data for the Crystal structure of ligand-free bifunctional folylpolyglutamate synthase/dihydrofolate synthase from yersinia pestis c092

First author:
B. Nocek
Gene name: folC
Resolution: 1.80 Å
R/Rfree: 0.17/0.19
Gene name: folC
Resolution: 1.80 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae soaked with octanoyl-CoA: conformational changes without clearly bound substrate

First author:
J. Hou
Resolution: 2.30 Å
R/Rfree: 0.20/0.23
Resolution: 2.30 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis

First author:
S.M. Anderson
Gene name: rocD
Resolution: 2.65 Å
R/Rfree: 0.20/0.23
Gene name: rocD
Resolution: 2.65 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with NAD and IMP

First author:
J. Osipiuk
Gene name: guaB
Resolution: 2.32 Å
R/Rfree: 0.20/0.27
Gene name: guaB
Resolution: 2.32 Å
R/Rfree: 0.20/0.27
X-ray diffraction data for the 3.0 Angstrom Crystal Structure of 3-dehydroquinate Synthase (AroB) from Francisella tularensis in Complex with NAD.

First author:
G. Minasov
Gene name: aroB
Resolution: 3.00 Å
R/Rfree: 0.16/0.19
Gene name: aroB
Resolution: 3.00 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal Structure of the Thiamin-bound form of Substrate-binding Protein of ABC Transporter from Clostridium difficile

First author:
Y. Kim
Resolution: 1.90 Å
R/Rfree: 0.16/0.20
Resolution: 1.90 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form

First author:
C. Chang
Gene name: folA
Resolution: 2.05 Å
R/Rfree: 0.17/0.22
Gene name: folA
Resolution: 2.05 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Inosine 5''-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with NAD and IMP.

First author:
N.Maltseva J.Osipiuk
Gene name: guaB
Resolution: 2.32 Å
R/Rfree: 0.20/0.27
Gene name: guaB
Resolution: 2.32 Å
R/Rfree: 0.20/0.27
X-ray diffraction data for the Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg in complex with D-Ala-D-Ala phosphinate analog

First author:
P.J. Stogios
Gene name: vanXYG
Resolution: 1.36 Å
R/Rfree: 0.16/0.18
Gene name: vanXYG
Resolution: 1.36 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the 1.7 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii with Tyrosine of Tag Bound to Active Site

First author:
G. Minasov
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the The crystal structure of azoreductase from Yersinia pestis CO92 in complex with FMN.

First author:
K. Tan
Gene name: acpD
Resolution: 1.45 Å
R/Rfree: 0.17/0.20
Gene name: acpD
Resolution: 1.45 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoD (3'-dephosphocoenzyme A)

First author:
K. Tan
Gene name: -
Resolution: 1.80 Å
R/Rfree: 0.16/0.21
Gene name: -
Resolution: 1.80 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP

First author:
K. Tan
Gene name: purK
Resolution: 2.06 Å
R/Rfree: 0.20/0.23
Gene name: purK
Resolution: 2.06 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the The crystal structure of the N-terminal domain of a MerR-like transcriptional regulator from Listeria monocytogenes EGD-e

First author:
K. Tan
Resolution: 1.87 Å
R/Rfree: 0.19/0.23
Resolution: 1.87 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the The crystal structure of thiol:disulfide interchange protein DsbC from Yersinia pestis CO92

First author:
K. Tan
Gene name: dsbC
Resolution: 2.15 Å
R/Rfree: 0.19/0.24
Gene name: dsbC
Resolution: 2.15 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a predicted ferric/iron (III) hydroxymate siderophore substrate binding protein from Bacillus anthracis

First author:
P.J. Stogios
Resolution: 2.47 Å
R/Rfree: 0.24/0.29
Resolution: 2.47 Å
R/Rfree: 0.24/0.29
X-ray diffraction data for the 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD


X-ray diffraction data for the 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis


X-ray diffraction data for the 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae


X-ray diffraction data for the 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.

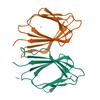
X-ray diffraction data for the Crystal structure of a fragment (1-405) of an elongation factor G from Vibrio vulnificus CMCP6


X-ray diffraction data for the Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with (S)-4-amino-5-fluoropentanoic acid


X-ray diffraction data for the 2.2 Angstrom Resolution Crystal Structure of P-Hydroxybenzoate Hydroxylase from Pseudomonas putida in Complex with FAD.


First author:
L.Shuvalova G.Minasov
Gene name: pobA
Resolution: 2.20 Å
R/Rfree: 0.17/0.22
Gene name: pobA
Resolution: 2.20 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Cycloalternan-forming enzyme from Listeria monocytogenes in complex with panose


X-ray diffraction data for the Cycloalternan-forming enzyme from Listeria monocytogenes in complex with maltopentaose


X-ray diffraction data for the 1.32 Angstrom Crystal Structure of Ybbr like Domain of lmo2119 Protein from Listeria monocytogenes.


X-ray diffraction data for the 1.95 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) E86A Mutant in Complex with Dehydroshikimate (Crystal Form #2)


X-ray diffraction data for the Structure of a putative reductase from Yersinia pestis


X-ray diffraction data for the Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with cycloalternan


X-ray diffraction data for the Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella enterica Typhimurium LT2 with Malonate and Boric Acid at the Active Site


X-ray diffraction data for the 2.0 Angstrom Resolution Crystal Structure of Transaldolase B (TalA) from Francisella tularensis in Covalent Complex with Fructose 6-Phosphate


X-ray diffraction data for the 1.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus (IDP00699) in complex with NAD+


X-ray diffraction data for the 2.50 angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with phosphoenolpyruvate


X-ray diffraction data for the Crystal structure of class D beta-lactamase from Sebaldella termitidis ATCC 33386


X-ray diffraction data for the 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica.


X-ray diffraction data for the 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD.


X-ray diffraction data for the 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium.


X-ray diffraction data for the Crystal structure of RNA-binding domain of nucleocapsid phosphoprotein from SARS CoV-2, monoclinic crystal form


First author:
C. Chang
Gene name: N
Resolution: 2.67 Å
R/Rfree: 0.20/0.25
Gene name: N
Resolution: 2.67 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Thiol-disulfide oxidoreductase TsdA, C129S mutant, from Corynebacterium diphtheriae


First author:
J. Osipiuk
Gene name: None
Resolution: 1.10 Å
R/Rfree: 0.12/0.13
Gene name: None
Resolution: 1.10 Å
R/Rfree: 0.12/0.13

First author:
M. Gucwa
Resolution: 2.40 Å
R/Rfree: 0.20/0.26
Resolution: 2.40 Å
R/Rfree: 0.20/0.26
X-ray diffraction data for the Crystal Structure of SrtB-anchored Collagen-binding Adhesin Fragment (residues 206-565) from Clostridioides difficile strain 630


First author:
G. Minasov
Resolution: 2.40 Å
R/Rfree: 0.17/0.22
Resolution: 2.40 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the 1.85 Angstrom Crystal Structure of O-succinylbenzoate Synthase from Salmonella typhimurium in Complex with Succinic Acid

First author:
G. Minasov
Gene name: menC
Resolution: 1.85 Å
R/Rfree: 0.16/0.18
Gene name: menC
Resolution: 1.85 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the 2.0 Angstrom Crystal Structure of an Acyl Carrier Protein S-malonyltransferase from Salmonella typhimurium.

First author:
G. Minasov
Gene name: fabD
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
Gene name: fabD
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Short chain dehydrogenase/reductase family oxidoreductase from Bacillus anthracis str. Ames Ancestor

First author:
E.V. Filippova
Resolution: 2.80 Å
R/Rfree: 0.20/0.26
Resolution: 2.80 Å
R/Rfree: 0.20/0.26
X-ray diffraction data for the 1.76A X-ray Crystal Structure of a Putative Phenazine Biosynthesis PhzC/PhzF Protein from Clostridium difficile (strain 630)

First author:
J.S. Brunzelle
Resolution: 1.76 Å
R/Rfree: 0.16/0.19
Resolution: 1.76 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Chaperone HscB from Vibrio cholerae

First author:
J. Osipiuk
Gene name: hscB
Resolution: 2.15 Å
R/Rfree: 0.22/0.26
Gene name: hscB
Resolution: 2.15 Å
R/Rfree: 0.22/0.26
X-ray diffraction data for the Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-bound

First author:
P.J. Stogios
Resolution: 2.03 Å
R/Rfree: 0.15/0.19
Resolution: 2.03 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal Structure of a Putative Succinate-Semialdehyde Dehydrogenase from Salmonella typhimurium LT2 with bound NAD

First author:
J.S. Brunzelle
Gene name: yneI
Resolution: 1.90 Å
R/Rfree: 0.16/0.19
Gene name: yneI
Resolution: 1.90 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, apo

First author:
P.J. Stogios
Gene name: ant4'-IIb
Resolution: 1.60 Å
R/Rfree: 0.17/0.19
Gene name: ant4'-IIb
Resolution: 1.60 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of putative dihydroneopterin aldolase (FolB) from Vibrio cholerae O1 biovar El Tor str. N16961

First author:
B. Nocek
Resolution: 1.95 Å
R/Rfree: 0.19/0.25
Resolution: 1.95 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the Crystal structure of C103A mutant of DJ-1 superfamily protein STM1931 from Salmonella typhimurium

First author:
I.A. Shumilin
Gene name: araH
Resolution: 2.20 Å
R/Rfree: 0.17/0.20
Gene name: araH
Resolution: 2.20 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile.

First author:
G. Minasov
Resolution: 2.00 Å
R/Rfree: 0.21/0.27
Resolution: 2.00 Å
R/Rfree: 0.21/0.27
X-ray diffraction data for the Crystal structure of carboxylesterase/phospholipase family protein from Francisella tularensis

First author:
E.V. Filippova
Resolution: 2.50 Å
R/Rfree: 0.20/0.26
Resolution: 2.50 Å
R/Rfree: 0.20/0.26
X-ray diffraction data for the Crystal Structure of Glutamate Racemase from Campylobacter jejuni subsp. jejuni

First author:
N. Maltseva
Gene name: murI
Resolution: 2.20 Å
R/Rfree: 0.19/0.24
Gene name: murI
Resolution: 2.20 Å
R/Rfree: 0.19/0.24