114 results
X-ray diffraction data for the HhaI endonuclease in Complex With DNA in space group P21 (pH 4.2)


First author:
J.R. Horton
Resolution: 1.16 Å
R/Rfree: 0.14/0.16
Resolution: 1.16 Å
R/Rfree: 0.14/0.16

First author:
C. Xu
Resolution: 1.80 Å
R/Rfree: 0.23/0.25
Resolution: 1.80 Å
R/Rfree: 0.23/0.25


First author:
C. Xu
Resolution: 2.10 Å
R/Rfree: 0.21/0.25
Resolution: 2.10 Å
R/Rfree: 0.21/0.25


First author:
J.C. Cofsky
Resolution: 1.64 Å
R/Rfree: 0.24/0.28
Resolution: 1.64 Å
R/Rfree: 0.24/0.28


First author:
C. Xu
Resolution: 1.85 Å
R/Rfree: 0.20/0.24
Resolution: 1.85 Å
R/Rfree: 0.20/0.24

First author:
C. Xu
Resolution: 2.85 Å
R/Rfree: 0.21/0.25
Resolution: 2.85 Å
R/Rfree: 0.21/0.25
X-ray diffraction data for the HhaI endonuclease in Complex with DNA at 1 Angstrom Resolution


First author:
J.R. Horton
Resolution: 1.00 Å
R/Rfree: 0.16/0.17
Resolution: 1.00 Å
R/Rfree: 0.16/0.17
X-ray diffraction data for the Crystal structure of Z-DNA in complex with putrescinium and potassium cations at ultrahigh-resolution

First author:
P. Drozdzal
Resolution: 0.60 Å
R/Rfree: 0.09/0.10
Resolution: 0.60 Å
R/Rfree: 0.09/0.10


First author:
M. Lei
Resolution: 2.30 Å
R/Rfree: 0.23/0.27
Resolution: 2.30 Å
R/Rfree: 0.23/0.27


First author:
Partnership for Stem Cell Biology Joint Center for Structural Genomics (JCSG)
Resolution: 2.77 Å
R/Rfree: 0.25/0.28
Resolution: 2.77 Å
R/Rfree: 0.25/0.28

First author:
C. Xu
Resolution: 2.10 Å
R/Rfree: 0.22/0.25
Resolution: 2.10 Å
R/Rfree: 0.22/0.25


First author:
M. Lei
Resolution: 1.84 Å
R/Rfree: 0.21/0.25
Resolution: 1.84 Å
R/Rfree: 0.21/0.25


First author:
K. Liu
Resolution: 2.05 Å
R/Rfree: 0.23/0.27
Resolution: 2.05 Å
R/Rfree: 0.23/0.27

First author:
C. Dong
Resolution: 1.70 Å
R/Rfree: 0.24/0.26
Resolution: 1.70 Å
R/Rfree: 0.24/0.26

First author:
K. Liu
Resolution: 2.30 Å
R/Rfree: 0.23/0.29
Resolution: 2.30 Å
R/Rfree: 0.23/0.29


First author:
L. Halabelian
Resolution: 2.20 Å
R/Rfree: 0.19/0.22
Resolution: 2.20 Å
R/Rfree: 0.19/0.22

First author:
L. Halabelian
Resolution: 2.60 Å
R/Rfree: 0.21/0.25
Resolution: 2.60 Å
R/Rfree: 0.21/0.25
X-ray diffraction data for the The crystal structure of the bacteriophage T4 MotA C-terminal domain in complex with dsDNA reveals a novel protein-DNA recognition motif


First author:
M.G. Cuypers
Resolution: 2.95 Å
R/Rfree: 0.22/0.25
Resolution: 2.95 Å
R/Rfree: 0.22/0.25


First author:
L. Halabelian
Resolution: 2.10 Å
R/Rfree: 0.21/0.24
Resolution: 2.10 Å
R/Rfree: 0.21/0.24


First author:
K. Liu
Resolution: 1.95 Å
R/Rfree: 0.22/0.26
Resolution: 1.95 Å
R/Rfree: 0.22/0.26


First author:
L. Halabelian
Resolution: 2.10 Å
R/Rfree: 0.21/0.24
Resolution: 2.10 Å
R/Rfree: 0.21/0.24


First author:
Y. Kim
Resolution: 2.50 Å
R/Rfree: 0.18/0.22
Resolution: 2.50 Å
R/Rfree: 0.18/0.22


First author:
Y. Kim
Resolution: 3.25 Å
R/Rfree: 0.24/0.28
Resolution: 3.25 Å
R/Rfree: 0.24/0.28


First author:
K.D. Koclega
Resolution: 3.20 Å
R/Rfree: 0.20/0.27
Resolution: 3.20 Å
R/Rfree: 0.20/0.27


First author:
Y. Kim
Resolution: 3.00 Å
R/Rfree: 0.17/0.21
Resolution: 3.00 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a DNA polymerase III subunit beta DnaN sliding clamp from Rickettsia typhi str. Wilmington


First author:
T.E. Edwards
Resolution: 2.00 Å
R/Rfree: 0.17/0.22
Resolution: 2.00 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of a DNA polymerase III subunit beta DnaN sliding clamp from Mycobacterium marinum


First author:
T.E. Edwards
Resolution: 2.45 Å
R/Rfree: 0.19/0.23
Resolution: 2.45 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of a DNA polymerase III subunit beta DnaN sliding clamp from Bartonella birtlesii LL-WM9


First author:
T.E. Edwards
Resolution: 2.45 Å
R/Rfree: 0.22/0.26
Resolution: 2.45 Å
R/Rfree: 0.22/0.26

First author:
K. Liu
Resolution: 2.01 Å
R/Rfree: 0.21/0.25
Resolution: 2.01 Å
R/Rfree: 0.21/0.25

First author:
K. Liu
Resolution: 1.90 Å
R/Rfree: 0.22/0.25
Resolution: 1.90 Å
R/Rfree: 0.22/0.25

First author:
K. Liu
Resolution: 2.65 Å
R/Rfree: 0.21/0.24
Resolution: 2.65 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Ultra-high resolution structure of d(CGCGCG)2 Z-DNA


First author:
K. Brzezinski
Resolution: 0.55 Å
R/Rfree: 0.08/0.09
Resolution: 0.55 Å
R/Rfree: 0.08/0.09
X-ray diffraction data for the Crystal Structure of dnaN DNA polymerase III beta subunit from Stenotrophomonas maltophilia K279a


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.15 Å
R/Rfree: 0.17/0.22
Resolution: 2.15 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of a DnaN sliding clamp (DNA polymerase III subunit beta) from Bartonella birtlesii bound to griselimycin


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.75 Å
R/Rfree: 0.18/0.21
Resolution: 1.75 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a DnaN sliding clamp (DNA polymerase III subunit beta) from Rickettsia rickettsii bound to griselimycin


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a DnaN sliding clamp (DNA polymerase III subunit beta) from Pseudomonas aeruginosa bound to griselimycin


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 3.05 Å
R/Rfree: 0.19/0.22
Resolution: 3.05 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Structure of a putative reductase from Yersinia pestis


X-ray diffraction data for the Mechanism of protease dependent DPC repair

First author:
F. Li
Resolution: 1.55 Å
R/Rfree: 0.16/0.19
Resolution: 1.55 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the C-Myc DNA binding protein complex


First author:
P. Aggarwal
Resolution: 2.57 Å
R/Rfree: 0.21/0.24
Resolution: 2.57 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of a DnaN sliding clamp DNA polymerase III subunit beta from Rickettsia bellii RML369-C


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID) Seattle Structural Genomics Center for Infectious Disease
Resolution: 2.35 Å
R/Rfree: 0.19/0.24
Resolution: 2.35 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the HhaI endonuclease in Complex with Iodine-Labelled DNA




First author:
S. Qin
Resolution: 1.80 Å
R/Rfree: 0.20/0.23
Resolution: 1.80 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the MeCP2 MBD in complex with DNA


First author:
M. Lei
Resolution: 1.80 Å
R/Rfree: 0.23/0.26
Resolution: 1.80 Å
R/Rfree: 0.23/0.26


X-ray diffraction data for the Full-length dimer of DNA-Damage Response Protein C from Deinococcus radiodurans - Crystal form xMJ7124


First author:
R. Szabla
Resolution: 4.28 Å
R/Rfree: 0.25/0.35
Resolution: 4.28 Å
R/Rfree: 0.25/0.35



First author:
M. Lei
Resolution: 2.30 Å
R/Rfree: 0.23/0.27
Resolution: 2.30 Å
R/Rfree: 0.23/0.27
X-ray diffraction data for the Complex of MBD1-MBD and methylated DNA

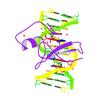
First author:
K. Liu
Resolution: 2.25 Å
R/Rfree: 0.24/0.27
Resolution: 2.25 Å
R/Rfree: 0.24/0.27
X-ray diffraction data for the Structure of apurinic/apyrimidinic DNA lyase TK0353 from Thermococcus kodakarensis (Iodide crystal form)

First author:
B.E. Eckenroth
Resolution: 2.20 Å
R/Rfree: 0.23/0.26
Resolution: 2.20 Å
R/Rfree: 0.23/0.26
X-ray diffraction data for the Structure of apurinic/apyrimidinic DNA lyase TK0353 from Thermococcus kodakarensis (Selenomethionine)


First author:
B.E. Eckenroth
Resolution: 1.98 Å
R/Rfree: 0.20/0.23
Resolution: 1.98 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Structure of apurinic/apyrimidinic DNA Lyase TK0353 from Thermococcus kodakarensis (Native Crystal Form)


First author:
B.E. Eckenroth
Resolution: 1.75 Å
R/Rfree: 0.19/0.22
Resolution: 1.75 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of DNA polymerase III subunit beta from Rickettsia conorii


First author:
K. Bowatte
Resolution: 1.70 Å
R/Rfree: 0.16/0.19
Resolution: 1.70 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the MeCP2 MBD in complex with DNA


First author:
M. Lei
Resolution: 1.65 Å
R/Rfree: 0.23/0.26
Resolution: 1.65 Å
R/Rfree: 0.23/0.26


First author:
X. Chao
Resolution: 2.05 Å
R/Rfree: 0.22/0.24
Resolution: 2.05 Å
R/Rfree: 0.22/0.24


First author:
X. Chao
Resolution: 2.25 Å
R/Rfree: 0.22/0.25
Resolution: 2.25 Å
R/Rfree: 0.22/0.25
X-ray diffraction data for the MBD2 in complex with methylated DNA

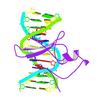
First author:
K. Liu
Resolution: 2.15 Å
R/Rfree: 0.21/0.23
Resolution: 2.15 Å
R/Rfree: 0.21/0.23
X-ray diffraction data for the Crystal Structure of the Transcription Factor AmrZ in Complex with the 18 Base Pair amrZ1 Binding Site


X-ray diffraction data for the Crystal Structure of DNase I Domain of Ribonuclease E from Vibrio cholerae


First author:
G. Minasov
Gene name: None
Resolution: 1.85 Å
R/Rfree: 0.21/0.24
Gene name: None
Resolution: 1.85 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of DNA polymerase III subunit beta from Rickettsia rickettsii


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Structure of DNA polymerase III subunit beta from Borrelia burgdorferi in complex with a natural product


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.05 Å
R/Rfree: 0.20/0.24
Resolution: 2.05 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12


First author:
G. Minasov
Resolution: 1.75 Å
R/Rfree: 0.16/0.18
Resolution: 1.75 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of a Heat shock protein (DnaJ) from Brucella melitensis


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: Q2YMC9
Resolution: 3.09 Å
R/Rfree: 0.24/0.29
Uniprot: Q2YMC9
Resolution: 3.09 Å
R/Rfree: 0.24/0.29
X-ray diffraction data for the DNA-binding protein HU from Bacillus anthracis

First author:
J. Osipiuk
Resolution: 2.48 Å
R/Rfree: 0.22/0.29
Resolution: 2.48 Å
R/Rfree: 0.22/0.29
X-ray diffraction data for the Crystal structure of a Putative bacterial DNA binding protein (BVU_2165) from Bacteroides vulgatus ATCC 8482 at 2.25 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.25 Å
R/Rfree: 0.17/0.21
Resolution: 2.25 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE DNA DAMAGE-INDUCIBLE PROTEIN (CHU_0679) FROM CYTOPHAGA HUTCHINSONII ATCC 33406 AT 1.50 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.16/0.19
Resolution: 1.50 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a Putative DNA replication regulator Hda (Sama_1916) from SHEWANELLA AMAZONENSIS SB2B at 3.00 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 3.00 Å
R/Rfree: 0.22/0.25
Resolution: 3.00 Å
R/Rfree: 0.22/0.25
X-ray diffraction data for the Structure of DNA polymerase III subunit beta from Rickettsia conorii in complex with a natural product


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.25 Å
R/Rfree: 0.18/0.23
Resolution: 2.25 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Structure of DNA polymerase III subunit beta from Rickettsia typhi in complex with a natural product


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of ferritin:DNA-binding protein DPS from Brucella Melitensis


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.70 Å
R/Rfree: 0.14/0.18
Resolution: 1.70 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10

First author:
K. Tan
Gene name: dps
Resolution: 2.75 Å
R/Rfree: 0.18/0.27
Gene name: dps
Resolution: 2.75 Å
R/Rfree: 0.18/0.27
X-ray diffraction data for the 2.17 Angstrom Crystal Structure of DNA-directed RNA Polymerase Subunit Alpha from Campylobacter jejuni.

First author:
G. Minasov
Gene name: rpoA
Resolution: 2.17 Å
R/Rfree: 0.17/0.23
Gene name: rpoA
Resolution: 2.17 Å
R/Rfree: 0.17/0.23
X-ray diffraction data for the DNA-binding transcriptional repressor AcrR from Salmonella typhimurium.

First author:
J. Osipiuk
Gene name: acrR
Resolution: 1.56 Å
R/Rfree: 0.15/0.20
Gene name: acrR
Resolution: 1.56 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the Crystal structure of a PAS and DNA binding domain containing protein (Caur_2278) from CHLOROFLEXUS AURANTIACUS J-10-FL at 2.30 A resolution


First author:
Q. Xu
Resolution: 2.30 Å
R/Rfree: 0.17/0.21
Resolution: 2.30 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a predicted dna-binding transcriptional regulator (saro_1072) from novosphingobium aromaticivorans dsm at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.17/0.20
Resolution: 2.10 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of the ATPase and transducer domains of DNA topoisomerase II from Balamuthia mandrillaris Lepto ID: CDC:V039: baboon/San Diego/1986


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.95 Å
R/Rfree: 0.17/0.20
Resolution: 1.95 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of DNA integrity scanning protein (DisA) from Mycobacterium tuberculosis in complex with cyclic di-AMP


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: P9WNW5
Resolution: 2.89 Å
R/Rfree: 0.20/0.25
Uniprot: P9WNW5
Resolution: 2.89 Å
R/Rfree: 0.20/0.25


First author:
Y. Hayashi
Resolution: 3.30 Å
R/Rfree: 0.24/0.27
Resolution: 3.30 Å
R/Rfree: 0.24/0.27
X-ray diffraction data for the Crystal structure of a DNA methyltransferase 1 associated protein 1 (DMAP1) from Homo sapiens at 1.45 A resolution


First author:
Partnership for T-Cell Biology Joint Center for Structural Genomics (JCSG)
Resolution: 1.45 Å
R/Rfree: 0.20/0.23
Resolution: 1.45 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae


First author:
G. Minasov
Resolution: 1.28 Å
R/Rfree: 0.14/0.18
Resolution: 1.28 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the Crystal structure of DNA integrity scanning protein DisA from Mycobacterium tuberculosis in complex with cyclic di-AMP and bromide

First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: P9WNW5
Resolution: 3.03 Å
R/Rfree: 0.23/0.27
Uniprot: P9WNW5
Resolution: 3.03 Å
R/Rfree: 0.23/0.27
X-ray diffraction data for the 1.5A Crystal Structure of a Putative Peptidase E Protein from Listeria monocytogenes EGD-e

First author:
J.S. Brunzelle
Resolution: 1.50 Å
R/Rfree: 0.17/0.22
Resolution: 1.50 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Structure of DNA polymerase III, beta subunit/ beta sliding clamp from Klebsiella pneumoniae, expressed with an N-terminal His-Smt3 fusion tag, in complex with Griselimycin


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.75 Å
R/Rfree: 0.19/0.21
Resolution: 1.75 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP


X-ray diffraction data for the Crystal structure of DNA polymerase III subunit beta from Mycobacterium marinum in complex with a natural product


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.10 Å
R/Rfree: 0.18/0.23
Resolution: 2.10 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase

First author:
S.M. Anderson
Resolution: 2.20 Å
R/Rfree: 0.15/0.19
Resolution: 2.20 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of putative transcriptional regulator containing a LuxR DNA binding domain (NP_811094.1) from Bacteroides thetaiotaomicron VPI-5482 at 2.04 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.04 Å
R/Rfree: 0.19/0.22
Resolution: 2.04 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida


X-ray diffraction data for the Crystal structure of Uracil-DNA glycosylase from Burkholderia thailandensis


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID) Seattle Structural Genomics Center for Infectious Disease
Uniprot: Q2SUH8
Resolution: 1.70 Å
R/Rfree: 0.16/0.20
Uniprot: Q2SUH8
Resolution: 1.70 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of a putative dna binding protein (ape_0880a) from aeropyrum pernix k1 at 2.21 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.21 Å
R/Rfree: 0.20/0.26
Resolution: 2.21 Å
R/Rfree: 0.20/0.26
X-ray diffraction data for the Crystal structure of DNA binding protein (YP_298823.1) from Ralstonia eutropha JMP134 at 1.92 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.92 Å
R/Rfree: 0.20/0.24
Resolution: 1.92 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of a histone family protein DNA-binding protein from Burkholderia ambifaria


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.85 Å
R/Rfree: 0.25/0.30
Resolution: 1.85 Å
R/Rfree: 0.25/0.30
X-ray diffraction data for the Crystal structure of predicted DNA-binding transcriptional regulator of TetR/AcrR family (NP_350189.1) from Clostridium acetobutylicum at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.20/0.23
Resolution: 2.10 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Crystal structure of a putative dna binding protein (bt_1116) from bacteroides thetaiotaomicron vpi-5482 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.14/0.17
Resolution: 1.50 Å
R/Rfree: 0.14/0.17


First author:
Partnership for Stem Cell Biology (STEMCELL) Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.18/0.23
Resolution: 1.85 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of Putative DNA-binding protein (YP_299413.1) from Ralstonia eutrophA JMP134 at 1.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.12/0.14
Resolution: 1.30 Å
R/Rfree: 0.12/0.14
X-ray diffraction data for the Crystal structure of a putative dna-binding protein (reut_b4095) from ralstonia eutropha jmp134 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.19/0.22
Resolution: 1.70 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Structure of mammalian NEIL2 from Monodelphis domestica in complex with THF-containing DNA


First author:
B.E. Eckenroth
Resolution: 2.08 Å
R/Rfree: 0.22/0.24
Resolution: 2.08 Å
R/Rfree: 0.22/0.24
X-ray diffraction data for the Crystal structure of a Putative DNA polymerase III beta subunit (EUBREC_0002; ERE_29750) from Eubacterium rectale ATCC 33656 at 2.26 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.26 Å
R/Rfree: 0.17/0.21
Resolution: 2.26 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE DNA-BINDING PROTEIN (CC_0111) FROM CAULOBACTER CRESCENTUS CB15 AT 1.62 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.62 Å
R/Rfree: 0.15/0.17
Resolution: 1.62 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE DNA DAMAGE-INDUCABLE (DINB) PROTEIN (BH3987) FROM BACILLUS HALODURANS AT 1.42 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.42 Å
R/Rfree: 0.16/0.18
Resolution: 1.42 Å
R/Rfree: 0.16/0.18