1469 results
X-ray diffraction data for the Crystal structure of a putative general stress family protein (xcc2264) from xanthomonas campestris pv. campestris at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.21/0.26
Resolution: 2.30 Å
R/Rfree: 0.21/0.26
X-ray diffraction data for the Crystal structure of a putative transcriptional regulator of the TETR family (SYN_02108) from Syntrophus aciditrophicus at 2.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.60 Å
R/Rfree: 0.24/0.26
Resolution: 2.60 Å
R/Rfree: 0.24/0.26
X-ray diffraction data for the Crystal structure of uncharacterized peroxidase-related protein (YP_614459.1) from Silicibacter sp. TM1040 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
Resolution: 1.70 Å
X-ray diffraction data for the Crystal structure of a putative plp-dependent beta-cystathionase (aecd, dip1736) from corynebacterium diphtheriae at 1.99 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.99 Å
R/Rfree: 0.15/0.18
Resolution: 1.99 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of Putative phosphoribosylformylglycinamidine cyclo-ligase (YP_676759.1) from CYTOPHAGA HUTCHINSONII ATCC 33406 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.16/0.19
Resolution: 1.50 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a putative nucleotide binding protein (tm0796) from Thermotoga maritima at 2.67 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.67 Å
R/Rfree: 0.21/0.24
Resolution: 2.67 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of a putative 6-phosphogluconolactonase (BACUNI_04672) from Bacteroides uniformis ATCC 8492 at 2.20 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.16/0.19
Resolution: 2.20 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE PHOSPHONOPYRUVATE HYDROLASE (MLL9387) FROM MESORHIZOBIUM LOTI MAFF303099 AT 2.15 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.16/0.20
Resolution: 2.15 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of putative glycoside hydrolase (YP_001304622.1) from Parabacteroides distasonis ATCC 8503 at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a putative antidote protein of plasmid maintenance system (npun_f2943) from nostoc punctiforme pcc 73102 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.17/0.20
Resolution: 1.60 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a putative iron-regulated protein A precursor (BDI_2603) from Parabacteroides distasonis ATCC 8503 at 2.30 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.16/0.19
Resolution: 2.30 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a protein with alpha-lytic protease prodomain-like fold (BDI_0842) from Parabacteroides distasonis ATCC 8503 at 1.40 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.13/0.17
Resolution: 1.40 Å
R/Rfree: 0.13/0.17
X-ray diffraction data for the Crystal structure of S-adenosylmethionine decarboxylase proenzyme (TM0655) from THERMOTOGA MARITIMA at 1.2 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.20 Å
Resolution: 1.20 Å
X-ray diffraction data for the Crystal structure of Putative metal-dependent phosphohydrolase (YP_926882.1) from Shewanella amazonensis SB2B at 2.06 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.06 Å
R/Rfree: 0.19/0.24
Resolution: 2.06 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of DNA polymerase III, beta chain (EC 2.7.7.7) (np_344555.1) from STREPTOCOCCUS PNEUMONIAE TIGR4 at 2.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.19/0.25
Resolution: 2.50 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the Crystal structure of Putative Acetyltransferase from the GNAT family (YP_497011.1) from NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.17/0.22
Resolution: 1.80 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of predicted HD superfamily hydrolase involved in NAD metabolism (NP_347894.1) from Clostridium acetobutylicum at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.18/0.22
Resolution: 1.50 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of p-coumaric acid decarboxylase (NP_786857.1) from Lactobacillus plantarum at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.18/0.21
Resolution: 1.70 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of an oligo-nucleotide binding protein (lpg1207) from Legionella pneumophila subsp. pneumophila str. philadelphia 1 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.20/0.23
Resolution: 2.00 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Crystal structure of Putative histidinol-phosphate aminotransferase (YP_050345.1) from Erwinia carotovora atroseptica SCRI1043 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.18/0.21
Resolution: 1.80 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a putative tetr/acrr family transcriptional regulator (saro_0558) from novosphingobium aromaticivorans dsm at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.18/0.21
Resolution: 1.80 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the CRYSTAL STRUCTURE OF A CADD-LIKE PROTEIN OF UNKNOWN FUNCTION (NPUN_F6505) FROM NOSTOC PUNCTIFORME PCC 73102 AT 1.35 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.35 Å
R/Rfree: 0.16/0.17
Resolution: 1.35 Å
R/Rfree: 0.16/0.17
X-ray diffraction data for the Crystal structure of Putative glycerophosphoryl diester phosphodiesterase (NP_812074.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.35 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.35 Å
R/Rfree: 0.15/0.17
Resolution: 1.35 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of a putative ca/calmodulin-dependent kinase ii association domain (exig_1688) from exiguobacterium sibiricum 255-15 at 2.59 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.59 Å
R/Rfree: 0.20/0.24
Resolution: 2.59 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of GCN5-related N-acetyltransferase-like protein (ZP_00874857) (ZP_00874857.1) from Streptococcus suis 89/1591 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.20/0.24
Resolution: 1.80 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of a HLA-B associated transcript 3 (BAT3) from Homo sapiens at 1.80 A resolution


First author:
Partnership for T-Cell Biology (TCELL) Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.18/0.24
Resolution: 1.80 Å
R/Rfree: 0.18/0.24
X-ray diffraction data for the Crystal structure of a prokaryotic domain of unknown function (duf849) with a tim barrel fold (bxe_c0966) from burkholderia xenovorans lb400 at 1.75 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.18/0.22
Resolution: 1.75 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of an IcmL-like type IV secretion system protein (lpg0120) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 at 2.65 A resolution

First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.65 Å
R/Rfree: 0.19/0.22
Resolution: 2.65 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of putative phosphosugar isomerase (YP_167080.1) from SILICIBACTER POMEROYI DSS-3 at 1.75 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.16/0.20
Resolution: 1.75 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of a PSPTO_0244 (Protein with unknown function which belongs to Pfam DUF861 family) from Pseudomonas syringae pv. tomato str. DC3000 at 1.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.12/0.15
Resolution: 1.30 Å
R/Rfree: 0.12/0.15
X-ray diffraction data for the Crystal structure of a putative lipoprotein from the DUF903 family (KPN_03160) from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 at 2.30 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.18/0.22
Resolution: 2.30 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a putative ribose 5-phosphate isomerase (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.81 Å
R/Rfree: 0.17/0.22
Resolution: 1.81 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of L-ALLO-threonine aldolase (tm1744) from Thermotoga maritima at 2.25 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.25 Å
R/Rfree: 0.14/0.22
Resolution: 2.25 Å
R/Rfree: 0.14/0.22
X-ray diffraction data for the Crystal structure of alkylhydroperoxidase AhpD core: uncharacterized peroxidase-related protein (YP_296737.1) from Ralstonia eutropha JMP134 at 2.15 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.21/0.26
Resolution: 2.15 Å
R/Rfree: 0.21/0.26
X-ray diffraction data for the Crystal structure of IMP dehydrogenase/GMP reductase-like protein (NP_599840.1) from Corynebacterium glutamicum ATCC 13032 Kitasato at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of Structural genomics protein of unknown function (NP_812590.1) from Bacteroides thetaiotaomicron VPI-5482 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.17/0.20
Resolution: 1.70 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of COG1633: Uncharacterized conserved protein (ZP_00055496.1) from Magnetospirillum magnetotacticum MS-1 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a Putative pilus assembly-related protein (BPSS2195) from Burkholderia pseudomallei K96243 at 2.50 A resolution (PSI Community Target, Shapiro L.)


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.19/0.26
Resolution: 2.50 Å
R/Rfree: 0.19/0.26
X-ray diffraction data for the Crystal structure of Putative glycerophosphoryl diester phosphodiesterase (17743486) from AGROBACTERIUM TUMEFACIENS str. C58 (Dupont) at 2.05 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.21/0.24
Resolution: 2.05 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE LACTONASE OF THE SMP-30/GLUCONOLACTONASE/LRE-LIKE REGION FAMILY (NPUN_F0524) FROM NOSTOC PUNCTIFORME PCC 73102 AT 1.90 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.17/0.22
Resolution: 1.90 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of putative cystathionine beta-lyase involved in aluminum resistance (NP_348457.1) from Clostridium acetobutylicum at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.15/0.19
Resolution: 2.00 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of Aspartate aminotransferase (tm1698) from Thermotoga maritima at 2.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.23/0.27
Resolution: 2.50 Å
R/Rfree: 0.23/0.27
X-ray diffraction data for the Crystal structure of TehB-like SAM-dependent methyltransferase (NP_600671.1) from Corynebacterium glutamicum ATCC 13032 Kitasato at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a putative phosphoenolpyruvate phosphonomutase (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.14/0.17
Resolution: 1.80 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of an alpha-lytic protease prodomain-like protein (DESPIG_01740) from Desulfovibrio piger ATCC 29098 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.20/0.22
Resolution: 1.80 Å
R/Rfree: 0.20/0.22
X-ray diffraction data for the Crystal structure of a predicted dna-binding transcriptional regulator (saro_1072) from novosphingobium aromaticivorans dsm at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.19/0.22
Resolution: 1.85 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE CYSTEINE DIOXYGENASE TYPE I (REUT_B5045) FROM RALSTONIA EUTROPHA JMP134 AT 1.84 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.84 Å
R/Rfree: 0.18/0.21
Resolution: 1.84 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of NTF2-like protein of unknown function MN2A_0505 from Prochlorococcus marinus (YP_291699.1) from Prochlorococcus sp. NATL2A at 1.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.17/0.18
Resolution: 1.40 Å
R/Rfree: 0.17/0.18
X-ray diffraction data for the Crystal structure of protein with a cupin-like fold and unknown function (YP_001165807.1) from NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 at 1.91 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.91 Å
R/Rfree: 0.17/0.21
Resolution: 1.91 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of predicted hydrolase of haloacid dehalogenase-like superfamily (NP_295428.1) from Deinococcus radiodurans at 1.66 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.66 Å
R/Rfree: 0.16/0.19
Resolution: 1.66 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a Cytoplasmic protein NCK2 (NCK2) from Homo sapiens at 2.20 A resolution

First author:
Partnership for T-Cell Biology (TCELL) Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.20/0.23
Resolution: 2.20 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE MODULATOR OF DNA GYRASE (BT3649) FROM BACTEROIDES THETAIOTAOMICRON VPI-5482 AT 1.75 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.18/0.20
Resolution: 1.75 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of a nipsnap family protein with unknown function (atu4242) from agrobacterium tumefaciens str. c58 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.15/0.18
Resolution: 1.50 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative polysaccharide deacetylase involved in o-antigen biosynthesis (wbms, bb0128) from bordetella bronchiseptica at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.16/0.18
Resolution: 1.90 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of Spore coat polysaccharide biosynthesis protein spsE (BSU37870) from Bacillus subtilis at 2.38 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.38 Å
R/Rfree: 0.19/0.24
Resolution: 2.38 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a putative phosphomethylpyrimidine kinase (BT_4458) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution (rhombohedral form)


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a putative s-adenosyl-l-methionine-dependent methyltransferase (mmp1179) from methanococcus maripaludis at 1.15 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.15 Å
R/Rfree: 0.12/0.14
Resolution: 1.15 Å
R/Rfree: 0.12/0.14
X-ray diffraction data for the Crystal structure of a putative gamma-carboxymuconolactone decarboxylase subunit (bxe_b0980) from burkholderia xenovorans lb400 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
Resolution: 1.70 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a putative metal binding protein RUMGNA_00854 (ZP_02040092.1) from Ruminococcus gnavus ATCC 29149 at 1.30 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.14/0.19
Resolution: 1.30 Å
R/Rfree: 0.14/0.19
X-ray diffraction data for the Crystal structure of putative acetyltransferase (YP_001815201.1) from EXIGUOBACTERIUM SP. 255-15 at 1.62 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.62 Å
R/Rfree: 0.18/0.20
Resolution: 1.62 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of FMN-dependent nitroreductase BF3017 from Bacteroides fragilis NCTC 9343 (YP_212631.1) from Bacteroides fragilis NCTC 9343 at 1.55 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.55 Å
R/Rfree: 0.16/0.19
Resolution: 1.55 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of Putative glycerophosphoryl diester phosphodiesterase (17743486) from AGROBACTERIUM TUMEFACIENS str. C58 (Dupont) at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.17/0.21
Resolution: 1.80 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a RNA binding motif protein 39 (RBM39) from Mus musculuS at 0.95 A resolution


First author:
Partnership for T-Cell Biology (TCELL) Joint Center for Structural Genomics (JCSG)
Resolution: 0.95 Å
R/Rfree: 0.12/0.13
Resolution: 0.95 Å
R/Rfree: 0.12/0.13
X-ray diffraction data for the Crystal structure of a putative nucleotidyltransferase (tm1012) from Thermotoga maritima at 1.35 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.20 Å
R/Rfree: 0.14/0.17
Resolution: 1.20 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of periplasmic sugar-binding protein (YP_001338366.1) from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 at 1.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.16/0.17
Resolution: 1.30 Å
R/Rfree: 0.16/0.17
X-ray diffraction data for the Crystal structure of Phosphoribosylamine--glycine ligase (TM1250) from Thermotoga maritima at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.21/0.26
Resolution: 2.30 Å
R/Rfree: 0.21/0.26
X-ray diffraction data for the CRYSTAL STRUCTURE OF a SAM dependent methyl-transferase type 12 family protein (ECA1738) FROM PECTOBACTERIUM ATROSEPTICUM SCRI1043 AT 1.74 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.74 Å
R/Rfree: 0.16/0.18
Resolution: 1.74 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the CRYSTAL STRUCTURE OF a YBJQ-LIKE FOLD PROTEIN OF UNKNOWN FUNCTION (BH3498) FROM BACILLUS HALODURANS AT 1.46 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.46 Å
R/Rfree: 0.15/0.18
Resolution: 1.46 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative acetyltransferase belonging to the gnat family (xcc2953) from xanthomonas campestris pv. campestris at 1.40 A resolution

First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.14/0.16
Resolution: 1.40 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of protein of unknown function with cystatin-like fold (YP_001022489.1) from METHYLOBIUM PETROLEOPHILUM PM1 at 1.45 A resolution

First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.45 Å
R/Rfree: 0.15/0.17
Resolution: 1.45 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of a SH3-SH2 domains of a lymphocyte-specific protein tyrosine kinase (LCK) from Homo sapiens at 2.36 A resolution


First author:
Partnership for T-Cell Biology JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.36 Å
R/Rfree: 0.19/0.25
Resolution: 2.36 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE OSMOTIC STRESS INDUCED AND DETOXIFICATION RESPONSE PROTEIN (PSYC_0566) FROM PSYCHROBACTER ARCTICUS 273-4 AT 2.15 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.20/0.24
Resolution: 1.95 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of a heat shock 70kDa protein 2 (HSPA2) from Homo sapiens at 1.80 A resolution


First author:
Partnership for T-Cell Biology (TCELL) Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.17/0.20
Resolution: 1.80 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a putative lipoprotein (CD630_1653) from Clostridium difficile 630 at 2.20 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.20/0.24
Resolution: 2.20 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE THIAMIN PHOSPHATE SYNTHASE (TM0723) FROM THERMOTOGA MARITIMA MSB8 AT 1.52 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.52 Å
R/Rfree: 0.14/0.16
Resolution: 1.52 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of Putative calcium/calmodulin-dependent protein kinase type II association domain (YP_315894.1) from THIOBACILLUS DENITRIFICANS ATCC 25259 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.01 Å
R/Rfree: 0.17/0.21
Resolution: 2.01 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of BH2720 (10175341) from Bacillus halodurans at 1.41 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.41 Å
R/Rfree: 0.17/0.19
Resolution: 1.41 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of the beta subunit of a putative aromatic-ring-hydroxylating dioxygenase (YP_001165631.1) from NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 at 1.75 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.17/0.21
Resolution: 1.75 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a RNA binding domain of polyadenylate-binding protein (PABPN1) from Homo sapiens at 1.95 A resolution


First author:
Partnership for T-Cell Biology JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.16/0.21
Resolution: 1.95 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of a branched chain amino acid ABC transporter periplasmic ligand-binding protein (Bxe_C0949) from BURKHOLDERIA XENOVORANS LB400 at 1.88 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.88 Å
R/Rfree: 0.19/0.23
Resolution: 1.88 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the CRYSTAL STRUCTURE OF A HYDROLASE FROM HALOACID DEHALOGENASE-LIKE FAMILY (SP_2064) FROM STREPTOCOCCUS PNEUMONIAE TIGR4 AT 2.10 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
Resolution: 2.10 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a 6-phosphogluconolactonase (Sbal_2240) from Shewanella baltica OS155 at 1.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.17/0.19
Resolution: 1.40 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF a pyridoxamine 5'-phosphate oxidase-related, FMN binding protein (NPUN_F5749) FROM NOSTOC PUNCTIFORME PCC 73102 AT 1.40 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.17/0.19
Resolution: 1.40 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE GENERAL STRESS PROTEIN 26 (JANN_0955) FROM JANNASCHIA SP. CCS1 AT 2.46 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.46 Å
R/Rfree: 0.23/0.28
Resolution: 2.46 Å
R/Rfree: 0.23/0.28
X-ray diffraction data for the Crystal structure of a Leucine binding protein LivK (TM1135) from Thermotoga maritima MSB8 at 1.90 A resolution


First author:
Joint center for structural genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.15/0.19
Resolution: 1.90 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a putative nadph-dependent oxidoreductase (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
Resolution: 1.70 Å
X-ray diffraction data for the Crystal structure of a putative cell surface protein (BACCAC_03700) from Bacteroides caccae ATCC 43185 at 2.07 A resolution

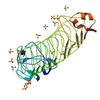
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.07 Å
R/Rfree: 0.18/0.21
Resolution: 2.07 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of Oligopeptide ABC transporter, periplasmic oligopeptide-binding (TM1223) from THERMOTOGA MARITIMA at 1.73 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.73 Å
R/Rfree: 0.15/0.18
Resolution: 1.73 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the CRYSTAL STRUCTURE OF a pyridoxamine 5'-phosphate oxidase-like family protein (NPUN_R6570) FROM NOSTOC PUNCTIFORME PCC 73102 AT 1.80 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.18/0.20
Resolution: 1.80 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of a putative gmp synthase subunit a protein (ta0944m) from thermoplasma acidophilum at 2.45 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.24 Å
R/Rfree: 0.18/0.25
Resolution: 2.24 Å
R/Rfree: 0.18/0.25
X-ray diffraction data for the Crystal structure of a Putative thua-like protein (PARMER_02418) from Parabacteroides merdae ATCC 43184 at 1.75 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.15/0.17
Resolution: 1.75 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of a diaminohydroxyphosphoribosylaminopyrimidine deaminase/ 5-amino-6-(5-phosphoribosylamino)uracil reductase (tm1828) from thermotoga maritima at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of putative glycosyl hydrolase, was Domain of Unknown Function (DUF1080) (YP_001302580.1) from Parabacteroides distasonis ATCC 8503 at 2.36 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.36 Å
R/Rfree: 0.20/0.25
Resolution: 2.36 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of sugar phosphate isomerase from a cupin superfamily SPO2919 from Silicibacter pomeroyi (YP_168127.1) from SILICIBACTER POMEROYI DSS-3 at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.20/0.25
Resolution: 2.30 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of a DUF4623 family protein (BACEGG_03550) from Bacteroides eggerthii DSM 20697 at 1.91 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.91 Å
R/Rfree: 0.15/0.19
Resolution: 1.91 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of Amino Transferase (RER070207001803) from Eubacterium rectale at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.17/0.22
Resolution: 2.10 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of Protein related to general stress protein 26(GS26) of B.subtilis (pyridoxinephosphate oxidase family) (NP_350077.1) from CLOSTRIDIUM ACETOBUTYLICUM at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a Deoxyribose-phosphate aldolase (TM_1559) from THERMOTOGA MARITIMA at 1.83 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.83 Å
R/Rfree: 0.15/0.19
Resolution: 1.83 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A C-TERMINAL FRAGMENT OF THE PUTATIVE FLAGELLAR MOTOR SWITCH PROTEIN FLIN (TM0680) FROM THERMOTOGA MARITIMA AT 1.85 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of the apo form of a 3alpha-hydroxysteroid dehydrogenase (BaiA2) associated with secondary bile acid synthesis from Clostridium scindens VPI12708 at 1.90 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.16/0.19
Resolution: 1.90 Å
R/Rfree: 0.16/0.19