1754 results
X-ray diffraction data for the Crystal structure of 2,5-diketo-D-gluconic acid reductase (TM1009) from Thermotoga maritima at 2.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.15/0.20
Resolution: 2.40 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the Crystal structure of putative TetR transcriptional regulator (YP_510936.1) from Jannaschia sp. CCS1 at 1.79 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.79 Å
R/Rfree: 0.21/0.24
Resolution: 1.79 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of a putative phosphoheptose isomerase (bh3325) from bacillus halodurans c-125 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.17/0.22
Resolution: 2.00 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of a cystatin-like fold protein (bxe_b1374) from burkholderia xenovorans lb400 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
Resolution: 2.00 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (TM1393) from Thermotoga maritima at 2.67 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.67 Å
R/Rfree: 0.20/0.25
Resolution: 2.67 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of a Calcium binding protein (BDI_1975) from Parabacteroides distasonis ATCC 8503 at 2.00 A resolution


First author:
Joint center for structural genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.19/0.22
Resolution: 2.00 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a Putative amino-acid aminotransferase (NP_104211.1) from Mesorhizobium loti at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.19/0.24
Resolution: 2.30 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE IRON-MOLYBDENUM COFACTOR (FEMO-CO) DINITROGENASE (TA1041M) FROM THERMOPLASMA ACIDOPHILUM DSM 1728 AT 1.30 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.16/0.18
Resolution: 1.30 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of a Putative flavoprotein (BACUNI_04544) from Bacteroides uniformis ATCC 8492 at 1.50 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of a putative secreted protein (BDI_1231) from Parabacteroides distasonis ATCC 8503 at 2.00 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.87 Å
R/Rfree: 0.20/0.24
Resolution: 1.87 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of a DUF4466 family protein (PARMER_03218) from Parabacteroides merdae ATCC 43184 at 2.00 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.17/0.22
Resolution: 2.00 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of a putative quercetin 2,3-dioxygenase (yxag, bsu39980) from bacillus subtilis at 2.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.60 Å
R/Rfree: 0.17/0.20
Resolution: 2.60 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of CheC-like superfamily protein (YP_001095400.1) from Shewanella SP. PV-4 at 2.13 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.13 Å
R/Rfree: 0.21/0.26
Resolution: 2.13 Å
R/Rfree: 0.21/0.26
X-ray diffraction data for the Crystal structure of a lipoprotein YedD (KPN_02420) from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 at 1.38 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.38 Å
R/Rfree: 0.15/0.18
Resolution: 1.38 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a PLP-dependent aminotransferase (ZP_03625122.1) from Streptococcus suis 89-1591 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.16/0.20
Resolution: 1.70 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of a Lipocalin-like protein (BACEGG_00036) from Bacteroides eggerthii DSM 20697 at 1.81 A resolution

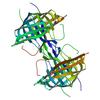
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.81 Å
R/Rfree: 0.17/0.20
Resolution: 1.81 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of GCN5-related N-acetyltransferase (YP_295895.1) from Ralstonia eutropha JMP134 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.14/0.17
Resolution: 1.80 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of a nitroreductase family protein (cd3355) from clostridium difficile 630 at 1.58 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.58 Å
R/Rfree: 0.18/0.22
Resolution: 1.58 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of phenolic acid decarboxylase (2635953) from Bacillus subtilis at 1.36 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.36 Å
R/Rfree: 0.13/0.15
Resolution: 1.36 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Crystal structure of a DUF4136 family protein (BT2437) from Bacteroides thetaiotaomicron VPI-5482 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.17/0.20
Resolution: 1.60 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a protein of unknown function (eca1910) from pectobacterium atrosepticum scri1043 at 2.20 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.19/0.22
Resolution: 2.20 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a putative cobalamin adenosyltransferase (bh1595) from bacillus halodurans c-125 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a Putative cell adhesion protein (BF1858) from Bacteroides fragilis NCTC 9343 at 2.33 A resolution


First author:
Joint center for structural genomics (JCSG)
Resolution: 2.33 Å
R/Rfree: 0.18/0.22
Resolution: 2.33 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF a putative 5-dehydro-2-deoxygluconokinase (IOLC) FROM BACILLUS HALODURANS C-125 AT 1.90 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.18/0.20
Resolution: 1.90 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of putative carboxypeptidase (YP_103406.1) from BURKHOLDERIA MALLEI ATCC 23344 at 2.49 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.49 Å
R/Rfree: 0.16/0.19
Resolution: 2.49 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a DUF1312 family protein (RUMGNA_02503) from Ruminococcus gnavus ATCC 29149 at 2.20 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.18/0.21
Resolution: 2.20 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.18/0.21
Resolution: 1.95 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a formyltetrahydrofolate deformylase (PP_0327) from PSEUDOMONAS PUTIDA KT2440 at 2.25 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.25 Å
R/Rfree: 0.22/0.27
Resolution: 2.25 Å
R/Rfree: 0.22/0.27
X-ray diffraction data for the Crystal structure of a DUF4783 family protein (BACUNI_04292) from Bacteroides uniformis ATCC 8492 at 1.27 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.27 Å
R/Rfree: 0.15/0.18
Resolution: 1.27 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative 2OG-Fe(II) oxygenase family protein (CC_0200) from CAULOBACTER CRESCENTUS at 1.44 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.44 Å
R/Rfree: 0.17/0.20
Resolution: 1.44 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a DUF5043 family protein (BACUNI_01052) from Bacteroides uniformis ATCC 8492 at 1.85 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.16/0.18
Resolution: 1.85 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of a putative GDSL-like lipase (BACOVA_00914) from Bacteroides ovatus ATCC 8483 at 1.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.13/0.16
Resolution: 1.40 Å
R/Rfree: 0.13/0.16
X-ray diffraction data for the Crystal structure of a putative alpha-L-fucosidase (BACOVA_04357) from Bacteroides ovatus ATCC 8483 at 1.59 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.59 Å
R/Rfree: 0.15/0.18
Resolution: 1.59 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative glycoside hydrolase (BVU_0247) from Bacteroides vulgatus ATCC 8482 at 2.46 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.46 Å
R/Rfree: 0.23/0.25
Resolution: 2.46 Å
R/Rfree: 0.23/0.25
X-ray diffraction data for the Crystal structure of a DUF2961 family protein (BACUNI_00161) from Bacteroides uniformis ATCC 8492 at 1.62 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.62 Å
R/Rfree: 0.14/0.16
Resolution: 1.62 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of a thioredoxine reductase (trxB) from Staphylococcus aureus subsp. aureus Mu50 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.16/0.17
Resolution: 1.80 Å
R/Rfree: 0.16/0.17
X-ray diffraction data for the Crystal structure of a DUF1329 family protein (DESPIG_00262) from Desulfovibrio piger ATCC 29098 at 1.75 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.21/0.25
Resolution: 1.75 Å
R/Rfree: 0.21/0.25
X-ray diffraction data for the Crystal structure of acetyltransferase of GNAT family (NP_373092.1) from STAPHYLOCOCCUS AUREUS MU50 at 2.20 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.17/0.22
Resolution: 2.20 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of a galactose mutarotase-like protein (CA_C0697) from CLOSTRIDIUM ACETOBUTYLICUM at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.14/0.18
Resolution: 1.80 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the Crystal structure of A Putative Acetyltransferase (NP_142035.1) from PYROCOCCUS HORIKOSHII at 2.25 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.25 Å
R/Rfree: 0.17/0.22
Resolution: 2.25 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of a putative dipeptidyl-peptidase VI (BACOVA_00612) from Bacteroides ovatus at 1.72 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.72 Å
R/Rfree: 0.14/0.17
Resolution: 1.72 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of a DUF4251 family protein (BACEGG_02002) from Bacteroides eggerthii DSM 20697 at 1.95 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.16/0.18
Resolution: 1.95 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of Geranyltranstransferase (EC 2.5.1.10) (tm0161) from THERMOTOGA MARITIMA at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.17/0.20
Resolution: 1.90 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a Putative thua-like protein (BACUNI_01602) from Bacteroides uniformis ATCC 8492 at 1.50 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.14/0.16
Resolution: 1.50 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of a putative GDSL-like lipase (PARMER_00689) from Parabacteroides merdae ATCC 43184 at 2.86 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.86 Å
R/Rfree: 0.18/0.23
Resolution: 2.86 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE LACCASE (YFIH) FROM ESCHERICHIA COLI AT 1.54 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.54 Å
R/Rfree: 0.14/0.17
Resolution: 1.54 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of dihydroorotase pyrC from Vibrio cholerae in complex with zinc at 1.95 A resolution.


First author:
J. Lipowska
Resolution: 1.95 Å
R/Rfree: 0.17/0.20
Resolution: 1.95 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of peptidase B from Yersinia pestis CO92 at 2.75 A resolution


First author:
M. Woinska
Gene name: pepB
Resolution: 2.75 Å
R/Rfree: 0.18/0.25
Gene name: pepB
Resolution: 2.75 Å
R/Rfree: 0.18/0.25
X-ray diffraction data for the 2.05 Angstrom Resolution Crystal Structure of Hypothetical Protein KP1_5497 from Klebsiella pneumoniae.


First author:
G. Minasov
Gene name: None
Resolution: 2.05 Å
R/Rfree: 0.20/0.23
Gene name: None
Resolution: 2.05 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the 1.9 Angstrom resolution crystal structure of uncharacterized protein lmo2446 from Listeria monocytogenes EGD-e


First author:
S.H. Light
Gene name: -
Resolution: 1.90 Å
R/Rfree: 0.15/0.18
Gene name: -
Resolution: 1.90 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the 2.65 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose reductase (rfbD) from Bacillus anthracis str. Ames in complex with NADP


First author:
A. Law
Gene name: rfbD
Resolution: 2.65 Å
R/Rfree: 0.22/0.26
Gene name: rfbD
Resolution: 2.65 Å
R/Rfree: 0.22/0.26
X-ray diffraction data for the Crystal structure of a tat-interacting protein homologue (htatip2, aw111545, cc3, tip30) from mus musculus at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.20/0.27
Resolution: 2.30 Å
R/Rfree: 0.20/0.27
X-ray diffraction data for the 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae.


First author:
G. Minasov
Gene name: wecB
Resolution: 2.35 Å
R/Rfree: 0.17/0.23
Gene name: wecB
Resolution: 2.35 Å
R/Rfree: 0.17/0.23
X-ray diffraction data for the Crystal structure of a putative cell adhesion protein (BACEGG_00536) from Bacteroides eggerthii DSM 20697 at 1.67 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.67 Å
R/Rfree: 0.16/0.19
Resolution: 1.67 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the 2.1 Angstrom Resolution Crystal Structure of Metallo-beta-lactamase from Staphylococcus aureus subsp. aureus COL


First author:
G. Minasov
Resolution: 2.10 Å
R/Rfree: 0.16/0.20
Resolution: 2.10 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the 1.88 Angstrom Resolution Crystal Structure of Hypothetical Protein jhp0584 from Helicobacter pylori.


First author:
G. Minasov
Resolution: 1.88 Å
R/Rfree: 0.17/0.21
Resolution: 1.88 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the CRYSTAL STRUCTURE OF A FMN-BINDING DOMAIN OF FLAVIN REDUCTASES-LIKE ENZYME (SBAL_0626) FROM SHEWANELLA BALTICA OS155 AT 1.50 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.13/0.15
Resolution: 1.50 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Crystal structure of Putative phosphatase (DUF442) (YP_001181608.1) from SHEWANELLA PUTREFACIENS CN-32 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative glycoside hydrolase (BT3745) from Bacteroides thetaiotaomicron VPI-5482 at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.18/0.21
Resolution: 2.10 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of Putative creatinine amidohydrolase (YP_211512.1) from Bacteroides fragilis NCTC 9343 at 2.11 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.11 Å
R/Rfree: 0.15/0.19
Resolution: 2.11 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a putative periplasmic proteins (BACEGG_01429) from Bacteroides eggerthii DSM 20697 at 2.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.20/0.21
Resolution: 2.40 Å
R/Rfree: 0.20/0.21
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE ENDORIBONUCLEASE (TM0215) FROM THERMOTOGA MARITIMA MSB8 AT 2.30 A RESOLUTION

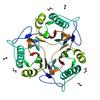
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.20/0.26
Resolution: 2.30 Å
R/Rfree: 0.20/0.26
X-ray diffraction data for the Crystal structure of Putative sugar binding protein (YP_001299726.1) from Bacteroides vulgatus ATCC 8482 at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.15/0.20
Resolution: 1.90 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the Crystal structure of a TetR family transcription regulator (Maqu_1417) from MARINOBACTER AQUAEOLEI VT8 at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.17/0.21
Resolution: 1.90 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of Putative carboxypeptidase A (YP_562911.1) from SHEWANELLA DENITRIFICANS OS-217 at 2.39 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.39 Å
R/Rfree: 0.18/0.23
Resolution: 2.39 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of a putative glycoside hydrolase (BDI_3141) from Parabacteroides distasonis ATCC 8503 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.15/0.17
Resolution: 1.70 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of inositol-3-phosphate synthase (ce21227) from Caenorhabditis elegans at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.20/0.27
Resolution: 2.30 Å
R/Rfree: 0.20/0.27
X-ray diffraction data for the Crystal structure of a putative dipeptidyl aminopeptidase IV (BACOVA_01349) from Bacteroides ovatus ATCC 8483 at 2.48 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.48 Å
R/Rfree: 0.17/0.21
Resolution: 2.48 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a putative cell wall hydrolase (CD630_03720) from Clostridium difficile 630 at 2.38 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.38 Å
R/Rfree: 0.18/0.21
Resolution: 2.38 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a putative kinase (caur_3907) from chloroflexus aurantiacus j-10-fl at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.18/0.22
Resolution: 1.70 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE NITROREDUCTASE IN COMPLEX WITH FMN (EXIG_2970) FROM EXIGUOBACTERIUM SIBIRICUM 255-15 AT 1.85 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
Resolution: 1.85 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of putative histidinol-phosphate aminotransferase (NP_281508.1) from Campylobacter jejuni at 2.01 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.01 Å
R/Rfree: 0.17/0.24
Resolution: 2.01 Å
R/Rfree: 0.17/0.24
X-ray diffraction data for the Crystal structure of a rmlc-like cupin family protein with a double-stranded beta-helix fold (mj0764) from methanocaldococcus jannaschii at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.15/0.20
Resolution: 1.70 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the Crystal structure of a putative anti-sigma factor (BDI_1681) from Parabacteroides distasonis ATCC 8503 at 1.65 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.65 Å
R/Rfree: 0.18/0.22
Resolution: 1.65 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a Putative transcriptional regulator (YP_001089212.1) from Clostridium difficile 630 at 1.86 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.86 Å
R/Rfree: 0.19/0.22
Resolution: 1.86 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a DUF4783 family protein (BF1468) from Bacteroides fragilis YCH46 at 1.55 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.55 Å
R/Rfree: 0.20/0.24
Resolution: 1.55 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of a cbs domain-containing protein in complex with amp (sso3205) from sulfolobus solfataricus at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.17/0.21
Resolution: 1.80 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of carbon-sulfur lyase involved in aluminum resistance (YP_878183.1) from CLOSTRIDIUM NOVYI NT at 2.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.90 Å
R/Rfree: 0.22/0.27
Resolution: 2.90 Å
R/Rfree: 0.22/0.27
X-ray diffraction data for the Crystal structure of a putative aminoglycoside phosphotransferase (YP_614837.1) from Silicibacter sp. TM1040 at 2.15 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.17/0.23
Resolution: 2.15 Å
R/Rfree: 0.17/0.23
X-ray diffraction data for the Crystal structure of a Putative bacterial DNA binding protein (BVU_2165) from Bacteroides vulgatus ATCC 8482 at 2.25 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.25 Å
R/Rfree: 0.17/0.21
Resolution: 2.25 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE DNA DAMAGE-INDUCIBLE PROTEIN (CHU_0679) FROM CYTOPHAGA HUTCHINSONII ATCC 33406 AT 1.50 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.16/0.19
Resolution: 1.50 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a protein with a cupin-like fold and unknown function (YP_400729.1) from Synechococcus SP. PCC 7942 (Elongatus) at 2.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.21/0.23
Resolution: 2.50 Å
R/Rfree: 0.21/0.23
X-ray diffraction data for the Crystal structure of a putative carbohydrate binding protein (BACOVA_03559) from Bacteroides ovatus ATCC 8483 at 1.50 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.16/0.18
Resolution: 1.50 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of Putative PLP-dependent aminotransferase (NP_978343.1) from Bacillus cereus ATCC 10987 at 2.19 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.19 Å
R/Rfree: 0.18/0.22
Resolution: 2.19 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a putative nuclease belonging to DUF820 (Ava_3926) from Anabaena variabilis ATCC 29413 at 1.96 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.96 Å
R/Rfree: 0.22/0.24
Resolution: 1.96 Å
R/Rfree: 0.22/0.24
X-ray diffraction data for the CRYSTAL STRUCTURE OF a transcriptional regulator of the tetR family (SHEW_3567) FROM SHEWANELLA LOIHICA PV-4 AT 2.05 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.05 Å
R/Rfree: 0.19/0.25
Resolution: 2.05 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the Crystal structure of a putative zn-dependent hydrolase of the metallo-beta-lactamase superfamily (tm0207) from thermotoga maritima at 2.00 A resolution

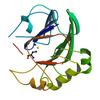
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a putative cell adhesion protein (LMOf2365_1307) from Listeria monocytogenes str. 4b F2365 at 1.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of Acyl-CoA hydrolase (15159470) from AGROBACTERIUM TUMEFACIENS at 2.65 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.23/0.24
Resolution: 2.50 Å
R/Rfree: 0.23/0.24
X-ray diffraction data for the Crystal structure of an Iron-containing alcohol dehydrogenase (Sden_2133) from Shewanella denitrificans OS-217 at 2.12 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.12 Å
R/Rfree: 0.18/0.20
Resolution: 2.12 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of aspartate aminotransferase (E.C. 2.6.1.1) (YP_194538.1) from Lactobacillus acidophilus NCFM at 2.15 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.18/0.24
Resolution: 2.15 Å
R/Rfree: 0.18/0.24
X-ray diffraction data for the Crystal structure of a putative zinc peptidase (NP_812461.1) from Bacteroides thetaiotaomicron VPI-5482 at 2.31 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.31 Å
R/Rfree: 0.19/0.25
Resolution: 2.31 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the Crystal structure of a two-domain protein containing dj-1/thij/pfpi-like and ferritin-like domains (ava_4496) from anabaena variabilis atcc 29413 at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.18/0.22
Resolution: 1.90 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of Putative phosphoheptose isomerase (YP_001815198.1) from Exiguobacterium sp. 255-15 at 1.95 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.16/0.19
Resolution: 1.95 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of putative acetyltransferase (YP_001815201.1) from EXIGUOBACTERIUM SP. 255-15 at 1.52 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.52 Å
R/Rfree: 0.16/0.18
Resolution: 1.52 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of a putative tetr-transcriptional regulator (sav143) from streptomyces avermitilis ma-4680 at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.19/0.23
Resolution: 2.10 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of a N-acetylmuramoyl-L-alanine amidase (BACUNI_02947) from Bacteroides uniformis ATCC 8492 at 1.15 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.15 Å
R/Rfree: 0.13/0.16
Resolution: 1.15 Å
R/Rfree: 0.13/0.16
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE SUPEROXIDE REDUCTASE (TM0658) FROM THERMOTOGA MARITIMA AT 2.00 A RESOLUTION

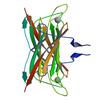
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.15/0.20
Resolution: 2.00 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the Crystal structure of a DUF4822 family protein (EF0375) from Enterococcus faecalis V583 at 1.85 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.15/0.18
Resolution: 1.85 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a formyltetrahydrofolate deformylase (PSPTO_4314) from Pseudomonas syringae pv. tomato str. DC3000 at 2.20 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.17/0.20
Resolution: 2.20 Å
R/Rfree: 0.17/0.20