1764 results
X-ray diffraction data for the Crystal structure of an aspartate transaminase (NCgl0237, Cgl0240) from CORYNEBACTERIUM GLUTAMICUM ATCC 13032 KITASATO at 1.25 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.25 Å
R/Rfree: 0.11/0.12
Resolution: 1.25 Å
R/Rfree: 0.11/0.12
X-ray diffraction data for the Crystal structure of the beta subunit of the benzoate 1,2-dioxygenase (benb, bmaa0186) from burkholderia mallei atcc 23344 at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.19/0.22
Resolution: 1.90 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of putative carboxymuconolactone decarboxylase (YP_555818.1) from Burkholderia xenovorans LB400 at 1.65 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.65 Å
R/Rfree: 0.15/0.18
Resolution: 1.65 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of Type I restriction enzyme StySJI M protein (NP_813429.1) from Bacteroides thetaiotaomicron VPI-5482 at 2.20 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.18/0.23
Resolution: 2.20 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of a tellurite resistance protein of cog3793 (npun_f6341) from nostoc punctiforme pcc 73102 at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.16/0.21
Resolution: 1.85 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of putative SnoaL-like polyketide cyclase (YP_001182657.1) from Shewanella putrefaciens CN-32 at 1.44 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.44 Å
R/Rfree: 0.15/0.17
Resolution: 1.44 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal structure of a pyridoxamine 5'-phosphate oxidase-related protein (ssuidraft_2804) from streptococcus suis 89/1591 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.19/0.23
Resolution: 2.00 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the CRYSTAL STRUCTURE OF AN UNCHARACTERIZED PROTEIN FROM DUF3069 FAMILY (SAMA_2622) FROM SHEWANELLA AMAZONENSIS SB2B AT 1.95 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.21/0.25
Resolution: 1.95 Å
R/Rfree: 0.21/0.25
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE CARBOHYDRATE KINASE (TM0922) FROM THERMOTOGA MARITIMA MSB8 AT 2.25 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.27 Å
R/Rfree: 0.16/0.22
Resolution: 2.27 Å
R/Rfree: 0.16/0.22
X-ray diffraction data for the Crystal structure of a ntf2-like protein of unknown function (sbal_0622) from shewanella baltica os155 at 1.75 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.15/0.19
Resolution: 1.75 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A FIC DOMAIN CONTAINING SIGNALING PROTEIN (BT_2513) FROM BACTEROIDES THETAIOTAOMICRON VPI-5482 AT 2.71 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.71 Å
R/Rfree: 0.22/0.25
Resolution: 2.71 Å
R/Rfree: 0.22/0.25
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE ANTIBIOTIC BIOSYNTHESIS MONOOXYGENASE (DR_2100) FROM DEINOCOCCUS RADIODURANS AT 1.40 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.13/0.16
Resolution: 1.40 Å
R/Rfree: 0.13/0.16
X-ray diffraction data for the Crystal structure of putative aminotransferase (MocR family) (YP_604413.1) from DEINOCOCCUS GEOTHERMALIS DSM 11300 at 2.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.60 Å
R/Rfree: 0.22/0.26
Resolution: 2.60 Å
R/Rfree: 0.22/0.26
X-ray diffraction data for the Crystal structure of a putative sulfurtransferase dsrE (Swol_2425) from Syntrophomonas wolfei str. Goettingen at 1.92 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.92 Å
R/Rfree: 0.19/0.22
Resolution: 1.92 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a SH2 domain of a megakaryocyte-associated tyrosine kinase (MATK) from Homo sapiens at 1.50 A resolution

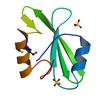
First author:
Partnership for T-Cell Biology (TCELL) Joint Center for Structural Genomics (JCSG)
Resolution: 1.50 Å
R/Rfree: 0.19/0.21
Resolution: 1.50 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the Crystal structure of 3-isopropylmalate dehydrogenase (TM0556) from Thermotoga maritima at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.19/0.22
Resolution: 1.90 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PHOSPHOSERINE AMINOTRANSFERASE SERC (CHU_0995) FROM CYTOPHAGA HUTCHINSONII ATCC 33406 AT 1.75 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.14/0.16
Resolution: 1.75 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of Putative sugar phosphate isomerase (AFE_0303) from Acidithiobacillus ferrooxidans ATCC 23270 at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.18/0.21
Resolution: 1.85 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of N-acetylglucosamine kinase (TM1224) from Thermotoga maritima at 2.46 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.46 Å
R/Rfree: 0.22/0.27
Resolution: 2.46 Å
R/Rfree: 0.22/0.27
X-ray diffraction data for the Crystal structure of a predicted zincin-like metalloprotease (acel_2062) from acidothermus cellulolyticus 11b at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.18/0.20
Resolution: 1.80 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of NTF2-like protein of unknown function (YP_680363.1) from CYTOPHAGA HUTCHINSONII ATCC 33406 at 1.45 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.45 Å
R/Rfree: 0.17/0.18
Resolution: 1.45 Å
R/Rfree: 0.17/0.18
X-ray diffraction data for the Crystal structure of transcriptional regulator of Crp/Fnr family (YP_604437.1) from DEINOCOCCUS GEOTHERMALIS DSM 11300 at 1.86 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.86 Å
R/Rfree: 0.21/0.25
Resolution: 1.86 Å
R/Rfree: 0.21/0.25
X-ray diffraction data for the Crystal structure of a sugar phosphate isomerase/epimerase (BDI_3400) from Parabacteroides distasonis ATCC 8503 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.14/0.16
Resolution: 1.70 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of a putative glycyl-glycine endopeptidase lytM (RUMGNA_02482) from Ruminococcus gnavus ATCC 29149 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the CRYSTAL STRUCTURE OF A putative acetyltransferase of the GNAT family (DDE_3044) FROM DESULFOVIBRIO DESULFURICANS SUBSP. AT 1.85 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.21/0.25
Resolution: 1.85 Å
R/Rfree: 0.21/0.25