6726 results
X-ray diffraction data for the Crystal structure of a Guanylate kinase from Cryptococcus neoformans var. grubii serotype A in complex with GDP and ADP


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of CdiA-CT/CdiI complex from Y. kristensenii 33638


First author:
K. Michalska
Resolution: 1.70 Å
R/Rfree: 0.17/0.20
Resolution: 1.70 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a putative pilus assembly protein (cpaE) from Burkholderia thailandensis E264 at 2.80 A resolution


First author:
Shapiro JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.80 Å
R/Rfree: 0.21/0.24
Resolution: 2.80 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of a Putative pilus assembly-related protein (BPSS2195) from Burkholderia pseudomallei K96243 at 2.50 A resolution (PSI Community Target, Shapiro L.)


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.19/0.26
Resolution: 2.50 Å
R/Rfree: 0.19/0.26
X-ray diffraction data for the Crystal structure of a CheY-like protein (tadZ) from Pseudomonas aeruginosa PAO1 at 2.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.20/0.23
Resolution: 2.40 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Crystal structure of a putative response regulator (Caur_3799) from Chloroflexus aurantiacus J-10-fl at 1.86 A resolution


First author:
Shapiro Joint center for structural genomics (JCSG)
Resolution: 1.86 Å
R/Rfree: 0.16/0.18
Resolution: 1.86 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of a putative CpaE2 pilus assembly protein (CpaE2) from Sinorhizobium meliloti 1021 at 2.72 A resolution (PSI Community Target, Shapiro)


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.72 Å
R/Rfree: 0.20/0.25
Resolution: 2.72 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of a Response regulator CheY-like protein (mll6475) from MESORHIZOBIUM LOTI at 2.02 A resolution


First author:
Shapiro Joint center for structural genomics (jcsg)
Resolution: 2.02 Å
R/Rfree: 0.20/0.22
Resolution: 2.02 Å
R/Rfree: 0.20/0.22
X-ray diffraction data for the Crystal structure of a FHA domain of kanadaptin (SLC4A1AP) from Homo sapiens at 1.55 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.55 Å
R/Rfree: 0.18/0.20
Resolution: 1.55 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of a centrosomal protein 170kDa, transcript variant beta (CEP170) from Homo sapiens at 2.15 A resolution (PSI Community Target, Sundstrom)


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.18/0.20
Resolution: 2.15 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of Histidine Phosphotransferase ShpA (NP_419930.1) from Caulobacter crescentus at 1.52 A resolution


First author:
Q. Xu
Resolution: 1.52 Å
R/Rfree: 0.17/0.20
Resolution: 1.52 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PROTEIN WITH UNKNOWN FUNCTION FROM DUF3598 FAMILY (NPUN_R4044) FROM NOSTOC PUNCTIFORME PCC 73102 AT 1.75 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.17/0.21
Resolution: 1.75 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of Methyltransferase FkbM (YP_546752.1) from Methylobacillus flagellatus KT at 2.20 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.17/0.22
Resolution: 2.15 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of a putative dioxygenase (npun_f5605) from nostoc punctiforme pcc 73102 at 2.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.60 Å
R/Rfree: 0.24/0.29
Resolution: 2.60 Å
R/Rfree: 0.24/0.29
X-ray diffraction data for the Crystal structure of uncharacterized peroxidase-related protein (YP_614459.1) from Silicibacter sp. TM1040 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
Resolution: 1.70 Å
X-ray diffraction data for the Crystal structure of an antioxidant defense protein (mlr4105) from mesorhizobium loti maff303099 at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.17/0.23
Resolution: 2.00 Å
R/Rfree: 0.17/0.23
X-ray diffraction data for the Crystal structure of a putative outer membrane chaperone (OmpH-like) (CC_1914) from Caulobacter crescentus CB15 at 2.83 A resolution (PSI Community Target, Shapiro)


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.83 Å
R/Rfree: 0.21/0.25
Resolution: 2.83 Å
R/Rfree: 0.21/0.25
X-ray diffraction data for the Crystal structure of a putative lipoprotein (CC_3750) from Caulobacter crescentus CB15 at 2.40 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.19/0.24
Resolution: 2.40 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a sensory box/GGDEF family protein (CC_0091) from Caulobacter crescentus CB15 at 1.40 A resolution (PSI Community Target, Shapiro)


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.40 Å
R/Rfree: 0.18/0.21
Resolution: 1.40 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of a putative dipeptidyl-peptidase VI (BT_1314) from Bacteroides thetaiotaomicron VPI-5482 at 1.75 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.15/0.18
Resolution: 1.75 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a putative dipeptidyl-peptidase VI (BT_1314) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.17/0.20
Resolution: 2.10 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a thioredoxine reductase (trxB) from Staphylococcus aureus subsp. aureus Mu50 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.16/0.17
Resolution: 1.80 Å
R/Rfree: 0.16/0.17
X-ray diffraction data for the Crystal structure of a fimbrilin (fimA) from Porphyromonas gingivalis W83 at 1.30 A resolution (PSI Community Target, Nakayama)


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.12/0.16
Resolution: 1.30 Å
R/Rfree: 0.12/0.16
X-ray diffraction data for the Crystal structure of a putative adhesin (BACOVA_02677) from Bacteroides ovatus ATCC 8483 at 3.00 A resolution (PSI Community Target, Nakayama)


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 3.00 Å
R/Rfree: 0.19/0.21
Resolution: 3.00 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the Crystal structure of a putative adhesin (BT2657) from Bacteroides thetaiotaomicron VPI-5482 at 2.20 A resolution (PSI Community Target, Nakayama)


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.17/0.22
Resolution: 2.20 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of Cryptosporidium parvum bromodomain cgd2_2690


First author:
L. Lin
Resolution: 1.38 Å
R/Rfree: 0.18/0.20
Resolution: 1.38 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of inositol-3-phosphate synthase (ce21227) from Caenorhabditis elegans at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.20/0.27
Resolution: 2.30 Å
R/Rfree: 0.20/0.27
X-ray diffraction data for the Chromo domain of human Chromodomain Protein, Y-Like 2


First author:
H. WU
Resolution: 2.00 Å
R/Rfree: 0.23/0.26
Resolution: 2.00 Å
R/Rfree: 0.23/0.26
X-ray diffraction data for the Crystal structure of a pilus assembly protein CpaE (CC_2943) from Caulobacter crescentus CB15 at 1.75 A resolution (PSI Community Target, Shapiro)


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.18/0.21
Resolution: 1.75 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the CBX3 chromo shadow domain in complex with histone H3 peptide


First author:
Y. Liu
Resolution: 1.60 Å
R/Rfree: 0.19/0.24
Resolution: 1.60 Å
R/Rfree: 0.19/0.24
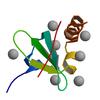
X-ray diffraction data for the chromo shadow domain of CBX1 in complex with a histone peptide

First author:
Y. Liu
Resolution: 1.90 Å
R/Rfree: 0.21/0.24
Resolution: 1.90 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of an acyl-ACP thioesterase (NP_810988.1) from Bacteroides thetaiotaomicron VPI-5482 at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.17/0.21
Resolution: 1.90 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal Structure of the complex between class B3 beta-lactamase BJP-1 and 4-nitrobenzene-sulfonamide


First author:
J.D. Docquier
Resolution: 1.33 Å
R/Rfree: 0.14/0.18
Resolution: 1.33 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the Crystal structure of a bile-acid 7-alpha dehydratase (CLOHIR_00079) from Clostridium hiranonis DSM 13275 at 1.89 A resolution with product added


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.89 Å
R/Rfree: 0.19/0.22
Resolution: 1.89 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a bile-acid 7-alpha dehydratase (CLOSCI_03134) from Clostridium scindens ATCC 35704 at 2.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.90 Å
R/Rfree: 0.18/0.22
Resolution: 2.90 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a bile-acid 7-alpha dehydratase (CLOHIR_00079) from Clostridium hiranonis DSM 13275 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of a bile-acid 7-alpha dehydratase (CLOHYLEM_06634) from Clostridium hylemonae DSM 15053 at 2.20 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.19/0.23
Resolution: 2.20 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of a bile acid-coenzyme A ligase (baiB) from Clostridium scindens (VPI 12708) at 2.19 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.19 Å
R/Rfree: 0.19/0.23
Resolution: 2.19 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of a 3alpha-hydroxysteroid dehydrogenase (BaiA2) associated with secondary bile acid synthesis from Clostridium scindens VPI12708 in complex with a putative NAD(+)-OH- adduct at 2.0 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.15/0.18
Resolution: 2.00 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of the apo form of a 3alpha-hydroxysteroid dehydrogenase (BaiA2) associated with secondary bile acid synthesis from Clostridium scindens VPI12708 at 1.90 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.16/0.19
Resolution: 1.90 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of AtS13 from Actinomadura melliaura


First author:
F. Wang
Resolution: 3.00 Å
R/Rfree: 0.19/0.22
Resolution: 3.00 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Structure of the universal stress protein from Archaeoglobus fulgidus in complex with dAMP


First author:
K.L. Tkaczuk
Resolution: 2.10 Å
R/Rfree: 0.20/0.24
Resolution: 2.10 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of putative acetolactate synthase- small subunit from Nitrosomonas europaea


First author:
J.J. Petkowski
Resolution: 2.50 Å
R/Rfree: 0.21/0.28
Resolution: 2.50 Å
R/Rfree: 0.21/0.28
X-ray diffraction data for the Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori cocrystallized with ATP


First author:
P.J. Porebski
Resolution: 2.80 Å
R/Rfree: 0.20/0.25
Resolution: 2.80 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold


First author:
S. Wang
Resolution: 2.40 Å
R/Rfree: 0.20/0.25
Resolution: 2.40 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis


First author:
A.M. Crowe
Resolution: 1.60 Å
R/Rfree: 0.16/0.20
Resolution: 1.60 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of PriA from Actinomyces urogenitalis


First author:
K. MICHALSKA
Resolution: 1.05 Å
R/Rfree: 0.12/0.14
Resolution: 1.05 Å
R/Rfree: 0.12/0.14
X-ray diffraction data for the Crystal structure of TrpF from Jonesia denitrificans


First author:
K. Michalska
Resolution: 1.09 Å
R/Rfree: 0.13/0.14
Resolution: 1.09 Å
R/Rfree: 0.13/0.14
X-ray diffraction data for the Crystal structure of HisAp form Arthrobacter aurescens


First author:
K. MICHALSKA
Resolution: 1.50 Å
R/Rfree: 0.14/0.17
Resolution: 1.50 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of HisAp from Streptomyces sviceus with degraded ProFAR


First author:
K. Michalska
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the CRYSTAL STRUCTURE OF HISAP FROM STREPTOMYCES SP. MG1


First author:
K. MICHALSKA
Resolution: 1.60 Å
R/Rfree: 0.16/0.19
Resolution: 1.60 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of HisAP from Streptomyces sp. Mg1


First author:
K. MICHALSKA
Resolution: 1.57 Å
R/Rfree: 0.15/0.18
Resolution: 1.57 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09


First author:
K. Michalska
Resolution: 2.80 Å
R/Rfree: 0.19/0.21
Resolution: 2.80 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the Crystal structure of aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon


First author:
K. Michalska
Resolution: 2.10 Å
R/Rfree: 0.21/0.23
Resolution: 2.10 Å
R/Rfree: 0.21/0.23
X-ray diffraction data for the PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20


First author:
K. Michalska
Resolution: 1.23 Å
R/Rfree: 0.14/0.17
Resolution: 1.23 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of CdiI from Burkholderia dolosa AUO158


First author:
K. Michalska
Resolution: 2.50 Å
R/Rfree: 0.19/0.21
Resolution: 2.50 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the Crystal structure of oxidoreductase, Gfo/Idh/MocA family from Streptococcus pneumoniae with reductive methylated Lysine


First author:
C. Chang
Resolution: 2.00 Å
R/Rfree: 0.15/0.20
Resolution: 2.00 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the Crystal structure of Tcur_1030 protein from Thermomonospora curvata


First author:
K. Michalska
Resolution: 2.11 Å
R/Rfree: 0.18/0.21
Resolution: 2.11 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata


First author:
J. Osipiuk
Resolution: 1.15 Å
R/Rfree: 0.12/0.15
Resolution: 1.15 Å
R/Rfree: 0.12/0.15
X-ray diffraction data for the ABC transporter substrate-binding protein from Sphaerobacter thermophilus


First author:
J. OSIPIUK
Resolution: 2.24 Å
R/Rfree: 0.17/0.22
Resolution: 2.24 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of universal stress protein E from Proteus mirabilis in complex with UDP-3-O-[(3R)-3-hydroxytetradecanoyl]-N-acetyl-alpha-glucosamine


First author:
I.A. Shumilin
Resolution: 1.80 Å
R/Rfree: 0.17/0.20
Resolution: 1.80 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Putative ribose ABC transporter, periplasmic solute-binding protein from Rhodobacter sphaeroides


First author:
J. Osipiuk
Resolution: 1.90 Å
R/Rfree: 0.13/0.14
Resolution: 1.90 Å
R/Rfree: 0.13/0.14
X-ray diffraction data for the L,D-transpeptidase from Mycobacterium smegmatis


First author:
J. Osipiuk
Resolution: 2.60 Å
R/Rfree: 0.20/0.24
Resolution: 2.60 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Transpeptidase domain of FtsI4 D,D-transpeptidase from Legionella pneumophila.


First author:
M. CUFF
Resolution: 2.10 Å
R/Rfree: 0.17/0.22
Resolution: 2.10 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the L,D-transpeptidase from Klebsiella pneumoniae


First author:
J. Osipiuk
Resolution: 1.90 Å
R/Rfree: 0.15/0.18
Resolution: 1.90 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of iron ABC transporter solute-binding protein from Eubacterium eligens


First author:
K. Michalska
Resolution: 1.85 Å
R/Rfree: 0.15/0.20
Resolution: 1.85 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the Phage-related protein DUF2815 from Enterococcus faecalis


First author:
J. Osipiuk
Resolution: 1.93 Å
R/Rfree: 0.21/0.25
Resolution: 1.93 Å
R/Rfree: 0.21/0.25
X-ray diffraction data for the ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii


First author:
J. Osipiuk
Resolution: 1.62 Å
R/Rfree: 0.15/0.19
Resolution: 1.62 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Structure of phage-related protein from Bacillus cereus ATCC 10987


First author:
E.V. Filippova
Resolution: 1.30 Å
R/Rfree: 0.12/0.16
Resolution: 1.30 Å
R/Rfree: 0.12/0.16
X-ray diffraction data for the Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin V


First author:
E.V. Filippova
Resolution: 1.85 Å
R/Rfree: 0.18/0.21
Resolution: 1.85 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469.


First author:
E.V. Filippova
Resolution: 1.92 Å
R/Rfree: 0.20/0.24
Resolution: 1.92 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Structural analysis of EspG2 glycosyltransferase


First author:
K. Michalska
Resolution: 2.70 Å
R/Rfree: 0.20/0.24
Resolution: 2.70 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the ent-Copalyl diphosphate synthase from Streptomyces platensis


First author:
J. Osipiuk
Resolution: 1.80 Å
R/Rfree: 0.16/0.19
Resolution: 1.80 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of cals8 from micromonospora Echinospora (P294S mutant)


First author:
K. Michalska
Resolution: 2.55 Å
R/Rfree: 0.18/0.23
Resolution: 2.55 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of LnmF protein from Streptomyces amphibiosporus


First author:
Archive MCSG
Resolution: 2.01 Å
R/Rfree: 0.18/0.23
Resolution: 2.01 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Beta-ketoacyl synthase domain (6118-6552) of polyketide synthase from Streptomyces albus


First author:
J. Osipiuk
Resolution: 2.12 Å
R/Rfree: 0.16/0.18
Resolution: 2.12 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Beta-ketoacyl synthase domain (4221-4804) of polyketide synthase from Streptomyces albus


First author:
J. Osipiuk
Resolution: 2.58 Å
R/Rfree: 0.18/0.23
Resolution: 2.58 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis


First author:
Y. Kim
Resolution: 2.78 Å
R/Rfree: 0.21/0.27
Resolution: 2.78 Å
R/Rfree: 0.21/0.27
X-ray diffraction data for the Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose)


First author:
K. Michalska
Resolution: 2.19 Å
R/Rfree: 0.16/0.21
Resolution: 2.19 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)


First author:
K. Michalska
Resolution: 2.00 Å
R/Rfree: 0.16/0.19
Resolution: 2.00 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal Structure of Conserved Uncharacterized Protein SgcJ from Streptomyces globisporus


First author:
Y. Kim
Resolution: 1.70 Å
R/Rfree: 0.17/0.20
Resolution: 1.70 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae


First author:
K. Michalska
Resolution: 2.28 Å
R/Rfree: 0.18/0.23
Resolution: 2.28 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of plu4264 protein from Photorhabdus luminescens


First author:
K. Michalska
Resolution: 1.35 Å
R/Rfree: 0.15/0.18
Resolution: 1.35 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment)


First author:
K. Michalska
Resolution: 1.98 Å
R/Rfree: 0.18/0.23
Resolution: 1.98 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of SSO0001


First author:
S. Lemak
Resolution: 2.35 Å
R/Rfree: 0.20/0.25
Resolution: 2.35 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Serine Protease HtrA3, mutationally inactivated


First author:
P. Glaza
Resolution: 3.27 Å
R/Rfree: 0.20/0.23
Resolution: 3.27 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL)


First author:
A.U. Singer
Resolution: 2.00 Å
R/Rfree: 0.17/0.23
Resolution: 2.00 Å
R/Rfree: 0.17/0.23
X-ray diffraction data for the Structure of the C-terminal domain of the type III effector Xcv3220 (XopL)


First author:
A.U. Singer
Resolution: 1.80 Å
R/Rfree: 0.15/0.20
Resolution: 1.80 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the Crystal structure of Legionella pneumophila Lpp1115 / KaiB


First author:
M. Loza-Correa
Resolution: 1.95 Å
R/Rfree: 0.19/0.25
Resolution: 1.95 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase


First author:
M. Rolando
Resolution: 2.85 Å
R/Rfree: 0.26/0.32
Resolution: 2.85 Å
R/Rfree: 0.26/0.32
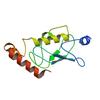
X-ray diffraction data for the Crystal structure of the complex between LubX/LegU2/Lpp2887 U-box 1 and Homo sapiens UBE2D2

First author:
A.T. Quaile
Resolution: 2.70 Å
R/Rfree: 0.17/0.22
Resolution: 2.70 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, Ile175Met mutant


First author:
A.T. Quaile
Resolution: 3.41 Å
R/Rfree: 0.18/0.22
Resolution: 3.41 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, wild-type


First author:
A.T. Quaile
Resolution: 2.88 Å
R/Rfree: 0.18/0.21
Resolution: 2.88 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of lpg1851 protein from Legionella pneumophila (putative T4SS effector)


First author:
K. Michalska
Resolution: 1.90 Å
R/Rfree: 0.18/0.23
Resolution: 1.90 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of lpg0076 protein from Legionella pneumophila


First author:
K. Michalska
Resolution: 2.05 Å
R/Rfree: 0.17/0.21
Resolution: 2.05 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of squalene synthase HpnC from Alicyclobacillus acidocaldarius


First author:
C. Chang
Resolution: 2.40 Å
R/Rfree: 0.20/0.25
Resolution: 2.40 Å
R/Rfree: 0.20/0.25
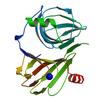
X-ray diffraction data for the Crystal structure of Cupin fold protein Sthe2323 from Sphaerobacter thermophilus

First author:
C. Chang
Resolution: 1.86 Å
R/Rfree: 0.16/0.21
Resolution: 1.86 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of member of Glyoxalase/bleomycin resistance protein/dioxygenase superfamily from Sphaerobacter thermophilus DSM 20745


First author:
B. Nocek
Resolution: 1.45 Å
R/Rfree: 0.14/0.18
Resolution: 1.45 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the Lmo2764 protein, a putative N-acetylmannosamine kinase, from Listeria monocytogenes


First author:
J. Osipiuk
Resolution: 1.64 Å
R/Rfree: 0.13/0.17
Resolution: 1.64 Å
R/Rfree: 0.13/0.17
X-ray diffraction data for the Crystal Structure of Transcriptional Antiterminator from Listeria monocytogenes EGD-e


First author:
Y. Kim
Resolution: 1.90 Å
R/Rfree: 0.17/0.22
Resolution: 1.90 Å
R/Rfree: 0.17/0.22