6637 results
X-ray diffraction data for the Crystal structure of Tryptophanyl-tRNA synthetase (EC 6.1.1.2) (Tryptophan-tRNA ligase)(TrpRS) (tm0492) from THERMOTOGA MARITIMA at 2.50 A resolution


First author:
G.W. Han
Resolution: 2.50 Å
R/Rfree: 0.19/0.26
Resolution: 2.50 Å
R/Rfree: 0.19/0.26
X-ray diffraction data for the Crystal structure of N5-glutamine methyltransferase, HemK(EC 2.1.1.-) (TM0488) from Thermotoga maritima at 2.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.80 Å
R/Rfree: 0.21/0.27
Resolution: 2.80 Å
R/Rfree: 0.21/0.27
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE THIAMINE BIOSYNTHESIS/SALVAGE PROTEIN (TM0486) FROM THERMOTOGA MARITIMA AT 1.80 A RESOLUTION


First author:
Z. Dermoun
Resolution: 1.80 Å
R/Rfree: 0.14/0.19
Resolution: 1.80 Å
R/Rfree: 0.14/0.19
X-ray diffraction data for the Crystal structure of the catalytic subunit of a phosphoribosylaminoimidazole mutase (tm0446) from thermotoga maritima at 1.77 A resolution


First author:
R. Schwarzenbacher
Resolution: 1.77 Å
R/Rfree: 0.15/0.18
Resolution: 1.77 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of Gluconate 5-dehydrogenase (TM0441) from Thermotoga maritima at 2.07 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.07 Å
R/Rfree: 0.15/0.18
Resolution: 2.07 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of Alcohol dehydrogenase (TM0436) from Thermotoga maritima at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.15/0.19
Resolution: 2.00 Å
R/Rfree: 0.15/0.19
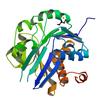
X-ray diffraction data for the Crystal structure of PfkB Carbohydrate kinase (TM0415) from Thermotoga maritima at 1.91 A resolution

First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.91 Å
R/Rfree: 0.17/0.22
Resolution: 1.91 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of glycerol-3-phosphate dehydrogenase (TM0378) from THERMOTOGA MARITIMA at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of Phospho-2-dehydro-3-deoxyheptonate aldolase (DAHP synthase) (TM0343) from Thermotoga Maritima at 1.92 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.92 Å
R/Rfree: 0.17/0.22
Resolution: 1.92 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of Orotidine 5'-phosphate decarboxylase (TM0332) from Thermotoga maritima at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of Oxidoreductase (TM0312) from Thermotoga maritima at 2.50 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.19/0.24
Resolution: 2.50 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of Transaldolase (EC 2.2.1.2) (TM0295) from Thermotoga maritima at 2.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.20/0.25
Resolution: 2.40 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of Activation (AdoMet binding) domain of Methionine synthase (TM0269) from Thermotoga maritima at 2.2 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.25/0.29
Resolution: 2.30 Å
R/Rfree: 0.25/0.29
X-ray diffraction data for the Crystal structure of DNA polymerase III, beta subunit (TM0262) from Thermotoga maritima at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.20/0.23
Resolution: 2.00 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE RNFG SUBUNIT OF ELECTRON TRANSPORT COMPLEX (TM0246) FROM THERMOTOGA MARITIMA AT 1.65 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.65 Å
R/Rfree: 0.18/0.21
Resolution: 1.65 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of Glycyl-tRNA synthetase alpha chain (TM0216) from Thermotoga maritima at 1.95 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.20/0.25
Resolution: 1.95 Å
R/Rfree: 0.20/0.25
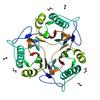
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE ENDORIBONUCLEASE (TM0215) FROM THERMOTOGA MARITIMA MSB8 AT 2.30 A RESOLUTION

First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.20/0.26
Resolution: 2.30 Å
R/Rfree: 0.20/0.26
X-ray diffraction data for the Crystal structure of Glycine cleavage system H protein (tm0212) from Thermotoga maritima at 1.65 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.65 Å
R/Rfree: 0.23/0.27
Resolution: 1.65 Å
R/Rfree: 0.23/0.27
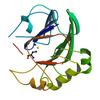
X-ray diffraction data for the Crystal structure of a putative zn-dependent hydrolase of the metallo-beta-lactamase superfamily (tm0207) from thermotoga maritima at 2.00 A resolution

First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a putative fe(iii) abc transporter (tm0189) from thermotoga maritima msb8 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.16/0.21
Resolution: 1.70 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of Acyl carrier protein (TM0175) from Thermotoga maritima at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.20/0.25
Resolution: 2.00 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of Folylpolyglutamate synthase (TM0166) from Thermotoga maritima at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.18/0.24
Resolution: 2.10 Å
R/Rfree: 0.18/0.24
X-ray diffraction data for the Crystal structure of Geranyltranstransferase (EC 2.5.1.10) (tm0161) from THERMOTOGA MARITIMA at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.17/0.20
Resolution: 1.90 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE XANTHOSINE TRIPHOSPHATE PYROPHOSPHATASE/HAM1 PROTEIN HOMOLOG (TM0159) FROM THERMOTOGA MARITIMA AT 1.78 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.78 Å
R/Rfree: 0.17/0.21
Resolution: 1.78 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of indole-3-glycerol phosphate synthase (TM0140) from Thermotoga maritima at 3.0 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 3.00 Å
R/Rfree: 0.24/0.30
Resolution: 3.00 Å
R/Rfree: 0.24/0.30