1764 results
X-ray diffraction data for the 2.65 Angstrom resolution crystal structure of an orotate phosphoribosyltransferase from Bacillus anthracis str. ''''Ames Ancestor'''' in complex with 5-phospho-alpha-D-ribosyl diphosphate (PRPP)

First author:
G.Minasov 'A.S.Halavaty
Gene name: pyrE
Resolution: 2.65 Å
R/Rfree: 0.20/0.24
Gene name: pyrE
Resolution: 2.65 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205

First author:
A.S. Halavaty
Gene name: metY
Resolution: 2.35 Å
R/Rfree: 0.20/0.25
Gene name: metY
Resolution: 2.35 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of Queuine tRNA-ribosyltransferase (EC 2.4.2.29) (tRNA-guanine (tm1561) from THERMOTOGA MARITIMA at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.19/0.23
Resolution: 1.90 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of a putative glyoxalase (NP_243026.1) from Bacillus halodurans at 2.10 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.10 Å
R/Rfree: 0.22/0.25
Resolution: 2.10 Å
R/Rfree: 0.22/0.25
X-ray diffraction data for the Crystal structure of a putative class i s-adenosylmethionine-dependent methyltransferase (lmo1582) from listeria monocytogenes at 2.20 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.17/0.21
Resolution: 2.20 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of a methyltransferase-like protein (spo2022) from silicibacter pomeroyi dss-3 at 1.80 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.80 Å
R/Rfree: 0.17/0.19
Resolution: 1.80 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the CRYSTAL STRUCTURE OF A PUTATIVE AROMATIC RING HYDROXYLASE (SARO_3538) FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM AT 1.75 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.17/0.20
Resolution: 1.75 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of SusD homolog (NP_813570.1) from Bacteroides thetaiotaomicron VPI-5482 at 1.70 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.16/0.19
Resolution: 1.70 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a fmn-binding protein (swol_0183) from syntrophomonas wolfei subsp. wolfei at 2.12 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.12 Å
R/Rfree: 0.22/0.25
Resolution: 2.12 Å
R/Rfree: 0.22/0.25
X-ray diffraction data for the Crystal structure of a duf1989 family protein (spo0365) from silicibacter pomeroyi dss-3 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.14/0.18
Resolution: 1.60 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the Crystal structure of a putative anti-sigma factor (BDI_1681) from Parabacteroides distasonis ATCC 8503 at 2.50 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.19/0.22
Resolution: 2.50 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of D-allose kinase (NP_418508.1) from ESCHERICHIA COLI K12 at 1.95 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.20/0.24
Resolution: 1.95 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of a putative lipoprotein (ycdA) from Bacillus subtilis subsp. subtilis str. 168 at 2.62 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.62 Å
R/Rfree: 0.21/0.24
Resolution: 2.62 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the 1.95 Angstrom Resolution Crystal Structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Yersinia pestis

First author:
G. Minasov
Resolution: 1.95 Å
R/Rfree: 0.16/0.20
Resolution: 1.95 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni.

First author:
G. Minasov
Resolution: 2.40 Å
R/Rfree: 0.18/0.23
Resolution: 2.40 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of a putative adhesin (PARMER_02777) from Parabacteroides merdae ATCC 43184 at 1.75 A resolution

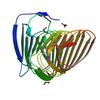
First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.75 Å
R/Rfree: 0.17/0.20
Resolution: 1.75 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a DUF3571 family protein (ABAYE3784) from Acinetobacter baumannii AYE at 1.95 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.95 Å
R/Rfree: 0.17/0.21
Resolution: 1.95 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the 2.06 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD-acetone

First author:
A.S. Halavaty
Resolution: 2.06 Å
R/Rfree: 0.16/0.21
Resolution: 2.06 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal structure of an uncharacterized protein (yobk, bsu18990) from bacillus subtilis at 1.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.14/0.18
Resolution: 1.30 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the Crystal structure of a porin-like protein (BACUNI_01323) from Bacteroides uniformis ATCC 8492 at 2.32 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.32 Å
R/Rfree: 0.17/0.19
Resolution: 2.32 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure of a fimbrilin (fimA) from Porphyromonas gingivalis W83 at 1.30 A resolution (PSI Community Target, Nakayama)


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.30 Å
R/Rfree: 0.12/0.16
Resolution: 1.30 Å
R/Rfree: 0.12/0.16
X-ray diffraction data for the Crystal structure of a DUF4847 family protein (BACEGG_01241) from Bacteroides eggerthii DSM 20697 at 2.23 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.23 Å
R/Rfree: 0.20/0.23
Resolution: 2.23 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the Crystal structure of a putative methyltransferase (bce_1332) from bacillus cereus atcc 10987 at 1.64 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.64 Å
R/Rfree: 0.17/0.20
Resolution: 1.64 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of a SusD superfamily protein (BF1802) from Bacteroides fragilis NCTC 9343 at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.15/0.19
Resolution: 1.90 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a DUF1541 family protein (ydhK) from Bacillus subtilis subsp. subtilis str. 168 at 2.00 A resolution

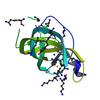
First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.18/0.26
Resolution: 2.00 Å
R/Rfree: 0.18/0.26