1469 results
X-ray diffraction data for the Crystal structure of a DUF2874 family protein (BACUNI_01296) from Bacteroides uniformis ATCC 8492 at 1.70 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.70 Å
R/Rfree: 0.19/0.21
Resolution: 1.70 Å
R/Rfree: 0.19/0.21
X-ray diffraction data for the Crystal structure of a putative monosaccharide binding protein (BACUNI_03039) from Bacteroides uniformis ATCC 8492 at 1.70 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 1.66 Å
R/Rfree: 0.16/0.18
Resolution: 1.66 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of an uncharacterized protein (bh2621) from bacillus halodurans at 1.55 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.55 Å
R/Rfree: 0.18/0.21
Resolution: 1.55 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the CRYSTAL STRUCTURE OF a DUF2118 family protein (MMP0046) FROM METHANOCOCCUS MARIPALUDIS AT 2.20 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.19/0.24
Resolution: 2.20 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a Putative lipoprotein (BF3042) from Bacteroides fragilis NCTC 9343 at 1.87 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.87 Å
R/Rfree: 0.19/0.22
Resolution: 1.87 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of an exotoxin 6 (SAV0422) from Staphylococcus aureus subsp. aureus Mu50 at 2.25 A resolution

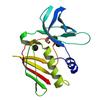
First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.25 Å
R/Rfree: 0.19/0.24
Resolution: 2.25 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a putative secreted protein (EUBREC_3654) from Eubacterium rectale at 2.43 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.43 Å
R/Rfree: 0.19/0.22
Resolution: 2.43 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of a transcriptional regulator, TetR family (BCE_2991) from Bacillus cereus ATCC 10987 at 2.50 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.50 Å
R/Rfree: 0.21/0.24
Resolution: 2.50 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of a putative adhesin (BACEGG_01763) from Bacteroides eggerthii DSM 20697 at 2.20 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.20 Å
R/Rfree: 0.21/0.24
Resolution: 2.20 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of a ferritin-like protein (pmt1231) from prochlorococcus marinus str. mit 9313 at 1.68 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.68 Å
R/Rfree: 0.16/0.20
Resolution: 1.68 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of an isochorismatase (PP1826) from Pseudomonas putida KT2440 at 1.60 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.60 Å
R/Rfree: 0.14/0.16
Resolution: 1.60 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal structure of a DUF4853 family protein (ACTODO_00621) from Actinomyces odontolyticus ATCC 17982 at 2.65 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.65 Å
R/Rfree: 0.23/0.26
Resolution: 2.65 Å
R/Rfree: 0.23/0.26
X-ray diffraction data for the Crystal structure of a DUF4398 family protein (PA2901) from Pseudomonas aeruginosa PAO1 at 2.47 A resolution


First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.47 Å
R/Rfree: 0.20/0.24
Resolution: 2.47 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal structure of Orotidine 5'-phosphate decarboxylase (TM0332) from Thermotoga maritima at 2.00 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
Resolution: 2.00 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the Crystal structure of a Histidine triad protein (Maqu_1709) from Marinobacter aquaeolei VT8 at 1.20 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.20 Å
R/Rfree: 0.13/0.15
Resolution: 1.20 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Crystal structure of a Pfam DUF1093 family protein (BSU39620) from Bacillus subtilis at 2.15 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.15 Å
R/Rfree: 0.18/0.22
Resolution: 2.15 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the CRYSTAL STRUCTURE OF a DUF416 family protein (SBAL_3149) FROM SHEWANELLA BALTICA OS155 AT 1.91 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.91 Å
R/Rfree: 0.17/0.22
Resolution: 1.91 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of a DUF4309 family protein (YjgB) from Bacillus subtilis subsp. subtilis str. 168 at 2.13 A resolution

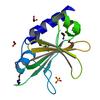
First author:
JOINT CENTER FOR STRUCTURAL GENOMICS (JCSG)
Resolution: 2.13 Å
R/Rfree: 0.19/0.23
Resolution: 2.13 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of a putative glucoamylase (BACUNI_03963) from Bacteroides uniformis ATCC 8492 at 2.01 A resolution


First author:
Joint center for structural genomics (JCSG)
Resolution: 2.01 Å
R/Rfree: 0.14/0.17
Resolution: 2.01 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of an iolb-like protein (stm4420) from salmonella typhimurium lt2 at 1.90 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.90 Å
R/Rfree: 0.16/0.18
Resolution: 1.90 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal structure of a putative sugar hydrolase (BACOVA_03189) from Bacteroides ovatus at 2.40 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.40 Å
R/Rfree: 0.17/0.22
Resolution: 2.40 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal structure of a dinb-like protein (yjoa, bsu12410) from bacillus subtilis at 2.30 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.30 Å
R/Rfree: 0.22/0.27
Resolution: 2.30 Å
R/Rfree: 0.22/0.27
X-ray diffraction data for the Crystal structure of a Sugar phosphate isomerase/epimerase (BDI_1903) from Parabacteroides distasonis ATCC 8503 at 1.85 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 1.85 Å
R/Rfree: 0.19/0.24
Resolution: 1.85 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the CRYSTAL STRUCTURE OF a DUF89 family protein (AF1104) FROM ARCHAEOGLOBUS FULGIDUS DSM 4304 AT 2.45 A RESOLUTION


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.45 Å
R/Rfree: 0.19/0.26
Resolution: 2.45 Å
R/Rfree: 0.19/0.26
X-ray diffraction data for the Crystal structure of a putative dna binding protein (ape_0880a) from aeropyrum pernix k1 at 2.21 A resolution


First author:
Joint Center for Structural Genomics (JCSG)
Resolution: 2.21 Å
R/Rfree: 0.20/0.26
Resolution: 2.21 Å
R/Rfree: 0.20/0.26