1117 results
X-ray diffraction data for the Succinyl-CoA synthase from Francisella tularensis, phosphorylated, in complex with CoA


First author:
J. Osipiuk
Resolution: 1.78 Å
R/Rfree: 0.18/0.21
Resolution: 1.78 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of thioredoxin-disulfide reductase from Vibrio vulnificus CMCP6 in complex with NADP and FAD


First author:
C. Chang
Gene name: trxB
Resolution: 2.60 Å
R/Rfree: 0.19/0.23
Gene name: trxB
Resolution: 2.60 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal Structure of Pantoate--Beta-Alanine Ligase from Campylobacter jejuni complexed with AMP and vitamin B5


First author:
Y. Kim
Gene name: panC
Resolution: 1.85 Å
R/Rfree: 0.17/0.20
Gene name: panC
Resolution: 1.85 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of azoreductase from Bacillus anthracis str. Sterne


First author:
E.V. Filippova
Resolution: 1.80 Å
R/Rfree: 0.19/0.23
Resolution: 1.80 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal Structure of Beta-lactamase/D-alanine Carboxypeptidase from Yersinia pestis complexed with citrate


First author:
Y. Kim
Gene name: ampH
Resolution: 1.50 Å
R/Rfree: 0.14/0.17
Gene name: ampH
Resolution: 1.50 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A


First author:
J. Hou
Resolution: 1.78 Å
R/Rfree: 0.17/0.20
Resolution: 1.78 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni subsp. jejuni NCTC 11168


First author:
Y. Kim
Gene name: dapA
Resolution: 1.84 Å
R/Rfree: 0.15/0.19
Gene name: dapA
Resolution: 1.84 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of cytidylyltransferase from Vibrio cholerae


First author:
J. Hattne
Resolution: 1.75 Å
R/Rfree: 0.20/0.23
Resolution: 1.75 Å
R/Rfree: 0.20/0.23
X-ray diffraction data for the 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis.


First author:
G. Minasov
Resolution: 2.30 Å
R/Rfree: 0.17/0.22
Resolution: 2.30 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the Crystal Structure of ATP-dependent Clp protease subunit P from Francisella tularensis


First author:
Y. Kim
Gene name: clpP
Resolution: 2.30 Å
R/Rfree: 0.19/0.23
Gene name: clpP
Resolution: 2.30 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the 1.88 Angstrom Resolution Crystal Structure of Hypothetical Protein jhp0584 from Helicobacter pylori.


First author:
G. Minasov
Resolution: 1.88 Å
R/Rfree: 0.17/0.21
Resolution: 1.88 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor'

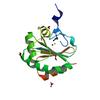
First author:
Y. Kim
Gene name: resA
Resolution: 1.35 Å
R/Rfree: 0.14/0.18
Gene name: resA
Resolution: 1.35 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH


First author:
J. Osipiuk
Gene name: dxr
Resolution: 2.60 Å
R/Rfree: 0.19/0.24
Gene name: dxr
Resolution: 2.60 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Metal ABC transporter from Listeria monocytogenes with cadmium


First author:
J. Osipiuk
Resolution: 1.72 Å
R/Rfree: 0.17/0.21
Resolution: 1.72 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames


First author:
N. Maltseva
Resolution: 1.60 Å
R/Rfree: 0.18/0.20
Resolution: 1.60 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68


First author:
Y. Kim
Gene name: guaB
Resolution: 2.60 Å
R/Rfree: 0.19/0.25
Gene name: guaB
Resolution: 2.60 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the Crystal Structure of Metallothiol Transferase from Bacillus anthracis str. Ames


First author:
N. Maltseva
Gene name: pal
Resolution: 1.52 Å
R/Rfree: 0.15/0.17
Gene name: pal
Resolution: 1.52 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni


First author:
Y. Kim
Gene name: guaB
Resolution: 2.12 Å
R/Rfree: 0.17/0.21
Gene name: guaB
Resolution: 2.12 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of Equine Serum Albumin complex with ibuprofen


First author:
M.P. Czub
Gene name: ALB
Resolution: 2.25 Å
R/Rfree: 0.19/0.24
Gene name: ALB
Resolution: 2.25 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant

First author:
J. Osipiuk
Gene name: orf1ab
Resolution: 1.60 Å
R/Rfree: 0.12/0.16
Gene name: orf1ab
Resolution: 1.60 Å
R/Rfree: 0.12/0.16
X-ray diffraction data for the Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with 3'-uridinemonophosphate


First author:
C. Chang
Resolution: 1.85 Å
R/Rfree: 0.17/0.19
Resolution: 1.85 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Putative ankyrin repeat domain-containing protein from Enterobacter cloacae

First author:
J. Osipiuk
Resolution: 1.60 Å
R/Rfree: 0.18/0.23
Resolution: 1.60 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of TrmD tRNA (guanine-N1)-methyltransferase from Corynebacterium diphtheriae in complex with SAH


First author:
K. Michalska
Gene name: trmD
Resolution: 1.35 Å
R/Rfree: 0.13/0.15
Gene name: trmD
Resolution: 1.35 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-2',3'-Vanadate


First author:
Y. Kim
Resolution: 2.25 Å
R/Rfree: 0.17/0.19
Resolution: 2.25 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-3',5'-Diphosphate

First author:
Y. Kim
Resolution: 2.40 Å
R/Rfree: 0.21/0.24
Resolution: 2.40 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal structure of Equine Serum Albumin in complex with Cobalt (II)


First author:
I.G. Shabalin
Gene name: ALB
Resolution: 2.70 Å
R/Rfree: 0.21/0.29
Gene name: ALB
Resolution: 2.70 Å
R/Rfree: 0.21/0.29
X-ray diffraction data for the 2.45 Angstrom Resolution Crystal Structure Thioredoxin Reductase from Francisella tularensis.


First author:
G. Minasov
Gene name: trxB
Resolution: 2.45 Å
R/Rfree: 0.22/0.25
Gene name: trxB
Resolution: 2.45 Å
R/Rfree: 0.22/0.25
X-ray diffraction data for the 1.73 Angstrom Resolution Crystal Structure of Dihydropteroate Synthase (folP-SMZ_B27) from Soil Uncultured Bacterium.


First author:
G. Minasov
Gene name: None
Resolution: 1.73 Å
R/Rfree: 0.16/0.19
Gene name: None
Resolution: 1.73 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073


First author:
J.S. Brunzelle
Gene name: pdhR
Resolution: 1.36 Å
R/Rfree: 0.18/0.21
Gene name: pdhR
Resolution: 1.36 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal structure of equine serum albumin in complex with testosterone


First author:
M.P. Czub
Gene name: ALB
Resolution: 2.15 Å
R/Rfree: 0.18/0.23
Gene name: ALB
Resolution: 2.15 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the 1.5 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) E86A Mutant


First author:
S.H. Light
Gene name: aroD
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
Gene name: aroD
Resolution: 1.50 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the 2.5 Angstrom Resolution Crystal Structure of Bifidobacterium longum Chorismate Synthase


First author:
S.H. Light
Gene name: aroC
Resolution: 2.50 Å
R/Rfree: 0.20/0.25
Gene name: aroC
Resolution: 2.50 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the 2.60 Angstrom Resolution Crystal Structure of Elongation Factor G 2 from Pseudomonas putida.


First author:
G. Minasov
Gene name: fusB
Resolution: 2.60 Å
R/Rfree: 0.22/0.26
Gene name: fusB
Resolution: 2.60 Å
R/Rfree: 0.22/0.26
X-ray diffraction data for the 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica.


First author:
G. Minasov
Resolution: 1.67 Å
R/Rfree: 0.17/0.20
Resolution: 1.67 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the High resolution structure of thioredoxin-disulfide reductase from Vibrio vulnificus CMCP6 in complex with NADP and FAD


First author:
C. Chang
Gene name: trxB
Resolution: 1.98 Å
R/Rfree: 0.15/0.20
Gene name: trxB
Resolution: 1.98 Å
R/Rfree: 0.15/0.20
X-ray diffraction data for the Crystal structure of dihydropteroate synthase from Klebsiella pneumoniae subsp.


First author:
C. Chang
Gene name: folP
Resolution: 2.60 Å
R/Rfree: 0.21/0.27
Gene name: folP
Resolution: 2.60 Å
R/Rfree: 0.21/0.27
X-ray diffraction data for the N-terminal domain of translation initiation factor IF-3 from Helicobacter pylori


First author:
J. Osipiuk
Gene name: infC
Resolution: 1.82 Å
R/Rfree: 0.18/0.21
Gene name: infC
Resolution: 1.82 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Crystal Structure of 2-C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase from Francisella tularensis


First author:
Y. Kim
Gene name: ispF
Resolution: 2.64 Å
R/Rfree: 0.17/0.23
Gene name: ispF
Resolution: 2.64 Å
R/Rfree: 0.17/0.23
X-ray diffraction data for the Structure of a phosphoglucosamine mutase from Francisella tularensis


First author:
J.S. Brunzelle
Gene name: mrsA
Resolution: 2.30 Å
R/Rfree: 0.19/0.24
Gene name: mrsA
Resolution: 2.30 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis


First author:
S.M. Anderson
Resolution: 1.75 Å
R/Rfree: 0.17/0.20
Resolution: 1.75 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of transcriptional regulator VanUg, Form I


First author:
P.J. Stogios
Gene name: vanUG
Resolution: 1.70 Å
R/Rfree: 0.16/0.20
Gene name: vanUG
Resolution: 1.70 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of a putative fumarylacetoacetate hydrolase family protein from Yersinia pestis CO92


First author:
B. Nocek
Resolution: 2.01 Å
R/Rfree: 0.16/0.21
Resolution: 2.01 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Structure of the YrdA ferripyochelin binding protein from Salmonella enterica


First author:
S.M. Anderson
Gene name: yrdA
Resolution: 1.20 Å
R/Rfree: 0.14/0.16
Gene name: yrdA
Resolution: 1.20 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the Crystal Structure of Amidotransferase HisH from Vibrio cholerae


First author:
N. Maltseva
Resolution: 1.91 Å
R/Rfree: 0.17/0.21
Resolution: 1.91 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis.

First author:
G. Minasov
Gene name: sipT
Resolution: 1.80 Å
R/Rfree: 0.17/0.20
Gene name: sipT
Resolution: 1.80 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase from Yersinia pestis CO92


First author:
B. Nocek
Gene name: ribB
Resolution: 1.95 Å
R/Rfree: 0.20/0.25
Gene name: ribB
Resolution: 1.95 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal Structure of Tryptophanyl-tRNA Synthetase from Campylobacter jejuni complexed with ADP and Tryptophane


First author:
Y. Kim
Gene name: trpS
Resolution: 2.15 Å
R/Rfree: 0.18/0.22
Gene name: trpS
Resolution: 2.15 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae.


First author:
G. Minasov
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
Resolution: 1.80 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of tryptophanyl-tRNA synthetase from Yersinia pestis CO92


First author:
B. Nocek
Gene name: trpS
Resolution: 1.95 Å
R/Rfree: 0.17/0.21
Gene name: trpS
Resolution: 1.95 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal Structure of Cytosol Aminopeptidase from Francisella tularensis


First author:
N. Maltseva
Gene name: pepA
Resolution: 2.70 Å
R/Rfree: 0.17/0.21
Gene name: pepA
Resolution: 2.70 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the 1.9 Angstrom Crystal Structure of Orotate Phosphoribosyltransferase (pyrE) Francisella tularensis.


First author:
G. Minasov
Gene name: pyrE
Resolution: 1.90 Å
R/Rfree: 0.19/0.24
Gene name: pyrE
Resolution: 1.90 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal structure of SP1627, a putative NADH-flavin reductase, from Streptococcus pneumoniae TIGR4


First author:
P.J. Stogios
Resolution: 2.40 Å
R/Rfree: 0.19/0.24
Resolution: 2.40 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the Crystal Structure of Aminoglycoside/Multidrug Efflux System AcrD from Salmonella typhimurium


First author:
Y. Kim
Gene name: acrD
Resolution: 3.00 Å
R/Rfree: 0.19/0.22
Gene name: acrD
Resolution: 3.00 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames


First author:
N. Maltseva
Resolution: 1.70 Å
R/Rfree: 0.17/0.19
Resolution: 1.70 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal structure putative autolysin from Listeria monocytogenes


First author:
C. Chang
Resolution: 2.15 Å
R/Rfree: 0.20/0.25
Resolution: 2.15 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of a probable undecaprenyl diphosphate synthase (uppS) from Campylobacter jejuni


First author:
B. Nocek
Gene name: uppS
Resolution: 2.46 Å
R/Rfree: 0.20/0.25
Gene name: uppS
Resolution: 2.46 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal Structure of Adenylate Kinase from Vibrio cholerae O1 biovar eltor


First author:
Y. Kim
Gene name: adk
Resolution: 2.00 Å
R/Rfree: 0.19/0.22
Gene name: adk
Resolution: 2.00 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose


First author:
Y. Kim
Gene name: mglB
Resolution: 1.32 Å
R/Rfree: 0.14/0.16
Gene name: mglB
Resolution: 1.32 Å
R/Rfree: 0.14/0.16
X-ray diffraction data for the 1.76 Angstrom Crystal Structure of GTP-binding Protein Der from Coxiella burnetii in Complex with GDP.


First author:
G. Minasov
Resolution: 1.76 Å
R/Rfree: 0.17/0.20
Resolution: 1.76 Å
R/Rfree: 0.17/0.20
X-ray diffraction data for the Crystal Structure of Ribonuclease Inhibitor Barstar from Salmonella Typhimurium


First author:
N. Maltseva
Gene name: yhcO
Resolution: 1.40 Å
R/Rfree: 0.14/0.18
Gene name: yhcO
Resolution: 1.40 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2


First author:
C. Chang
Gene name: N
Resolution: 1.70 Å
R/Rfree: 0.16/0.21
Gene name: N
Resolution: 1.70 Å
R/Rfree: 0.16/0.21
X-ray diffraction data for the Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose


First author:
K. Michalska
Gene name: orf1ab
Resolution: 1.50 Å
R/Rfree: 0.15/0.17
Gene name: orf1ab
Resolution: 1.50 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES


First author:
K. Michalska
Gene name: orf1ab
Resolution: 1.07 Å
R/Rfree: 0.13/0.15
Gene name: orf1ab
Resolution: 1.07 Å
R/Rfree: 0.13/0.15
X-ray diffraction data for the Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with potential repurposing drug Tipiracil


First author:
Y. Kim
Resolution: 1.85 Å
R/Rfree: 0.17/0.19
Resolution: 1.85 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with the Product Nucleotide GpU.


First author:
Y. Kim
Resolution: 1.97 Å
R/Rfree: 0.16/0.18
Resolution: 1.97 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate


First author:
Y. Kim
Resolution: 1.82 Å
R/Rfree: 0.17/0.19
Resolution: 1.82 Å
R/Rfree: 0.17/0.19
X-ray diffraction data for the The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with a Lys48-linked di-ubiquitin


First author:
J. Osipiuk
Resolution: 1.88 Å
R/Rfree: 0.19/0.23
Resolution: 1.88 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the The crystal structure of Papain-Like Protease of SARS CoV-2, C111S/D286N mutant, in complex with a Lys48-linked di-ubiquitin


First author:
J. Osipiuk
Resolution: 1.45 Å
R/Rfree: 0.14/0.18
Resolution: 1.45 Å
R/Rfree: 0.14/0.18
X-ray diffraction data for the Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 1

First author:
M. Gucwa
Gene name: ALB
Resolution: 2.40 Å
R/Rfree: 0.18/0.26
Gene name: ALB
Resolution: 2.40 Å
R/Rfree: 0.18/0.26
X-ray diffraction data for the Crystal structure of the ENTH domain of ENT2 from Candida albicans


First author:
P.J. Stogios
Gene name: None
Resolution: 1.83 Å
R/Rfree: 0.16/0.19
Gene name: None
Resolution: 1.83 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the 1.7 Angstrom Crystal Structure of of Putative Modulator of Drug Activity (apo- form) from Yersinia pestis CO92


First author:
G. Minasov
Gene name: mdaB
Resolution: 1.70 Å
R/Rfree: 0.15/0.17
Gene name: mdaB
Resolution: 1.70 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM


First author:
C. Chang
Gene name: panC
Resolution: 2.40 Å
R/Rfree: 0.18/0.24
Gene name: panC
Resolution: 2.40 Å
R/Rfree: 0.18/0.24
X-ray diffraction data for the Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis


First author:
Y. Kim
Resolution: 2.00 Å
R/Rfree: 0.20/0.22
Resolution: 2.00 Å
R/Rfree: 0.20/0.22
X-ray diffraction data for the Crystal structure of the putative tol-pal system-associated acyl-CoA thioesterase from Pseudomonas aeruginosa PAO1


First author:
D. Borek
Resolution: 1.90 Å
R/Rfree: 0.22/0.28
Resolution: 1.90 Å
R/Rfree: 0.22/0.28
X-ray diffraction data for the 2.1 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae in Complex with L-Malate


First author:
G. Minasov
Gene name: mdh
Resolution: 2.10 Å
R/Rfree: 0.17/0.22
Gene name: mdh
Resolution: 2.10 Å
R/Rfree: 0.17/0.22
X-ray diffraction data for the X-ray structure of a putative triosephosphate isomerase from Toxoplasma gondii ME49


First author:
E.V. Filippova
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.


First author:
G. Minasov
Resolution: 2.61 Å
R/Rfree: 0.23/0.28
Resolution: 2.61 Å
R/Rfree: 0.23/0.28
X-ray diffraction data for the 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid.


First author:
G. Minasov
Resolution: 1.36 Å
R/Rfree: 0.14/0.17
Resolution: 1.36 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD


First author:
G. Minasov
Resolution: 1.88 Å
R/Rfree: 0.14/0.17
Resolution: 1.88 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the 1.55 Angstrom Resolution Crystal Structure of Glutathione Reductase from Yersinia pestis in Complex with FAD


First author:
G. Minasov
Resolution: 1.55 Å
R/Rfree: 0.15/0.17
Resolution: 1.55 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni.


First author:
G. Minasov
Gene name: murA
Resolution: 2.46 Å
R/Rfree: 0.22/0.28
Gene name: murA
Resolution: 2.46 Å
R/Rfree: 0.22/0.28
X-ray diffraction data for the 2.3 Angstrom Resolution Crystal Structure of Glutathione Reductase from Vibrio parahaemolyticus in Complex with FAD.


First author:
G. Minasov
Resolution: 2.31 Å
R/Rfree: 0.20/0.25
Resolution: 2.31 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus


First author:
G. Minasov
Gene name: pbp2X
Resolution: 1.95 Å
R/Rfree: 0.17/0.21
Gene name: pbp2X
Resolution: 1.95 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae


First author:
G. Minasov
Gene name: ponA
Resolution: 2.61 Å
R/Rfree: 0.18/0.22
Gene name: ponA
Resolution: 2.61 Å
R/Rfree: 0.18/0.22
X-ray diffraction data for the 2.2 Angstrom Resolution Crystal Structure of P-Hydroxybenzoate Hydroxylase from Pseudomonas putida in Complex with FAD.


First author:
G. Minasov
Gene name: pobA
Resolution: 2.20 Å
R/Rfree: 0.18/0.23
Gene name: pobA
Resolution: 2.20 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the 1.5 Angstrom Resolution Crystal Structure of NAD-Dependent Epimerase from Klebsiella pneumoniae in Complex with NAD.


First author:
G. Minasov
Gene name: uge
Resolution: 1.50 Å
R/Rfree: 0.18/0.21
Gene name: uge
Resolution: 1.50 Å
R/Rfree: 0.18/0.21
X-ray diffraction data for the Cycloalternan-forming enzyme from Listeria monocytogenes in complex with cycloalternan


First author:
S.H. Light
Gene name: -
Resolution: 1.77 Å
R/Rfree: 0.14/0.17
Gene name: -
Resolution: 1.77 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the 1.50 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Shikimate


First author:
S.H. Light
Gene name: aroD
Resolution: 1.50 Å
R/Rfree: 0.16/0.18
Gene name: aroD
Resolution: 1.50 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the Cycloalternan-forming enzyme from Listeria monocytogenes in complex with pentasaccharide substrate


First author:
S.H. Light
Gene name: -
Resolution: 1.70 Å
R/Rfree: 0.16/0.19
Gene name: -
Resolution: 1.70 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of Listeria monocytogenes InlP


First author:
S. Nocadello
Resolution: 1.40 Å
R/Rfree: 0.16/0.18
Resolution: 1.40 Å
R/Rfree: 0.16/0.18
X-ray diffraction data for the 1.78 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Quinate


First author:
S.H. Light
Gene name: aroD
Resolution: 1.78 Å
R/Rfree: 0.16/0.19
Gene name: aroD
Resolution: 1.78 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of a putative C-S lyase from Bacillus anthracis


First author:
S.M. Anderson
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
Resolution: 2.00 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein


First author:
J.S. Brunzelle
Gene name: None
Resolution: 1.55 Å
R/Rfree: 0.17/0.21
Gene name: None
Resolution: 1.55 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the 1.55 Angstrom Resolution Crystal Structure of 6-phosphogluconolactonase from Klebsiella pneumoniae


First author:
G. Minasov
Gene name: pgl
Resolution: 1.55 Å
R/Rfree: 0.13/0.16
Gene name: pgl
Resolution: 1.55 Å
R/Rfree: 0.13/0.16
X-ray diffraction data for the 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD and Mn2+.


First author:
G. Minasov
Gene name: aglB
Resolution: 2.25 Å
R/Rfree: 0.16/0.22
Gene name: aglB
Resolution: 2.25 Å
R/Rfree: 0.16/0.22
X-ray diffraction data for the 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae.


First author:
G. Minasov
Resolution: 1.93 Å
R/Rfree: 0.19/0.24
Resolution: 1.93 Å
R/Rfree: 0.19/0.24
X-ray diffraction data for the 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168


First author:
G. Minasov
Gene name: argI
Resolution: 1.70 Å
R/Rfree: 0.14/0.17
Gene name: argI
Resolution: 1.70 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD.


First author:
G. Minasov
Gene name: aglB
Resolution: 2.25 Å
R/Rfree: 0.15/0.19
Gene name: aglB
Resolution: 2.25 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of uncharacterized protein ECL_02694


First author:
C. Chang
Resolution: 1.45 Å
R/Rfree: 0.15/0.18
Resolution: 1.45 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal Structure of the H233A mutant of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with preQ0


First author:
Y. Kim
Resolution: 1.62 Å
R/Rfree: 0.15/0.18
Resolution: 1.62 Å
R/Rfree: 0.15/0.18