1376 results
X-ray diffraction data for the Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD bound, orthorhombic form)


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.81 Å
R/Rfree: 0.17/0.21
Resolution: 1.81 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (Apo, hexagonal form)


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.10 Å
R/Rfree: 0.19/0.25
Resolution: 2.10 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD bound, No sulfate hexagonal form)
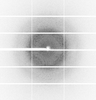

First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.90 Å
R/Rfree: 0.18/0.23
Resolution: 1.90 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal Structure of Betaine aldehyde dehydrogenase (BetB) from Klebsiella aerogenes (Lattice Translocation Disorder)


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.80 Å
R/Rfree: 0.17/0.21
Resolution: 1.80 Å
R/Rfree: 0.17/0.21
X-ray diffraction data for the Crystal structure of outer membrane lipoprotein carrier protein (LolA) from Borrelia burgdorferi (steric acid bound)


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.80 Å
R/Rfree: 0.20/0.24
Resolution: 1.80 Å
R/Rfree: 0.20/0.24
X-ray diffraction data for the Crystal Structure of UDP-N-acetylmuramoylalanine--D-glutamate ligase (MurD) from E. coli in complex with UMA and two inhibitor A19 molecules
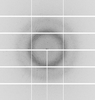

First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: P14900
Resolution: 1.23 Å
R/Rfree: 0.12/0.14
Uniprot: P14900
Resolution: 1.23 Å
R/Rfree: 0.12/0.14
X-ray diffraction data for the Crystal structure of outer membrane lipoprotein carrier protein (LolA) from Francisella philomiragia (Orthorhombic P Form)


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: B0TX44
Resolution: 1.70 Å
R/Rfree: 0.18/0.23
Uniprot: B0TX44
Resolution: 1.70 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal structure of Acetyl-CoA Synthetase in Complex with Adenosine-5'-propylphosphate from Coccidioides posadasii C735


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: C5PGB4
Resolution: 2.15 Å
R/Rfree: 0.15/0.19
Uniprot: C5PGB4
Resolution: 2.15 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of Ornithine carbamoyltransferase from Burkholderia xenovorans in complex with phosphono carbamate
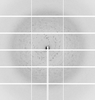

First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: Q13H08
Resolution: 2.67 Å
R/Rfree: 0.19/0.25
Uniprot: Q13H08
Resolution: 2.67 Å
R/Rfree: 0.19/0.25
X-ray diffraction data for the Crystal structure of DNA integrity scanning protein (DisA) from Mycobacterium tuberculosis in complex with cyclic di-AMP


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: P9WNW5
Resolution: 2.89 Å
R/Rfree: 0.20/0.25
Uniprot: P9WNW5
Resolution: 2.89 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of Cysteinyl-tRNA synthetase (CysRS) from Plasmodium falciparum in complex with cysteinyl-AMP


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: Q8IJP3
Resolution: 2.22 Å
R/Rfree: 0.18/0.20
Uniprot: Q8IJP3
Resolution: 2.22 Å
R/Rfree: 0.18/0.20
X-ray diffraction data for the Crystal structure of Apo Cysteinyl-tRNA synthetase (CysRS) from Plasmodium falciparum (Orthrhombic P form)


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: Q8IJP3
Resolution: 3.06 Å
R/Rfree: 0.23/0.29
Uniprot: Q8IJP3
Resolution: 3.06 Å
R/Rfree: 0.23/0.29
X-ray diffraction data for the Crystal structure of Cysteinyl-tRNA synthetase (CysRS) from Plasmodium falciparum in complex with AMP and Cysteine


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: Q8IJP3
Resolution: 2.34 Å
R/Rfree: 0.19/0.22
Uniprot: Q8IJP3
Resolution: 2.34 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of Cysteinyl-tRNA synthetase (CysRS) from Plasmodium falciparum in complex with Cysteine


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: Q8IJP3
Resolution: 2.16 Å
R/Rfree: 0.19/0.22
Uniprot: Q8IJP3
Resolution: 2.16 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of Cysteinyl-tRNA synthetase (CysRS) from Plasmodium falciparum in complex with ATP (long soak)


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: Q8IJP3
Resolution: 2.80 Å
R/Rfree: 0.20/0.22
Uniprot: Q8IJP3
Resolution: 2.80 Å
R/Rfree: 0.20/0.22