1376 results
X-ray diffraction data for the Crystal Structure of calcium-dependent protein kinase 1 (CDPK1) from Cryptosporidium parvum (AMP/Mg bound)


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: A3FQ16
Resolution: 2.12 Å
R/Rfree: 0.21/0.24
Uniprot: A3FQ16
Resolution: 2.12 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal Structure of UDP-N-acetylmuramoylalanine--D-glutamate ligase (MurD) from E. coli in complex with UMA and AMP-PNP


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: P14900
Resolution: 1.45 Å
R/Rfree: 0.15/0.16
Uniprot: P14900
Resolution: 1.45 Å
R/Rfree: 0.15/0.16
X-ray diffraction data for the Crystal structure of Ornithine carbamoyltransferase from Burkholderia xenovorans


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: Q13H08
Resolution: 2.55 Å
R/Rfree: 0.19/0.23
Uniprot: Q13H08
Resolution: 2.55 Å
R/Rfree: 0.19/0.23
X-ray diffraction data for the Crystal structure of glutamate dehydrogenase from Babesia microti in complex with NADP


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: A0A0K3AUK4
Resolution: 2.28 Å
R/Rfree: 0.19/0.22
Uniprot: A0A0K3AUK4
Resolution: 2.28 Å
R/Rfree: 0.19/0.22
X-ray diffraction data for the Crystal structure of Cysteinyl-tRNA synthetase (CysRS) from Plasmodium falciparum (Hexagonal P form)


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.87 Å
R/Rfree: 0.22/0.24
Resolution: 2.87 Å
R/Rfree: 0.22/0.24
X-ray diffraction data for the Crystal structure of a calcium bound C2 domain containing protein from Trichomonas vaginalis


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: A2EZR3
Resolution: 1.40 Å
R/Rfree: 0.14/0.17
Uniprot: A2EZR3
Resolution: 1.40 Å
R/Rfree: 0.14/0.17
X-ray diffraction data for the Crystal Structure of 6,7-dimethyl-8-ribityllumazine synthase from Bordetella pertussis

First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Uniprot: Q7VTN4
Resolution: 2.34 Å
R/Rfree: 0.21/0.26
Uniprot: Q7VTN4
Resolution: 2.34 Å
R/Rfree: 0.21/0.26
X-ray diffraction data for the Crystal structure of a peptide deformylase from Burkholderia ambifaria


First author:
Abendroth Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
Resolution: 1.60 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of phosphoprotein/Protein P/Protein M1 residues 69-297 from Rabies virus reveals degradation to C-terminal domain only

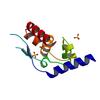
First author:
T.E. Edwards
Resolution: 2.20 Å
R/Rfree: 0.17/0.23
Resolution: 2.20 Å
R/Rfree: 0.17/0.23
X-ray diffraction data for the Crystal structure of nucleoside diphosphate kinase from Giardia lamblia featuring a disordered dinucleotide binding site


First author:
T.E. Edwards
Resolution: 2.65 Å
R/Rfree: 0.23/0.27
Resolution: 2.65 Å
R/Rfree: 0.23/0.27
X-ray diffraction data for the Crystal structure of Glutamate-1-semialdehyde 2,1- aminomutase (GSA) from Pseudomonas aeruginosa


First author:
Delker Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.75 Å
R/Rfree: 0.15/0.18
Resolution: 1.75 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal Structure of Prolyl-tRNA Synthetase from Cryptosporidium parvum complexed with L-Proline and AMP


First author:
Dranow Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.80 Å
R/Rfree: 0.21/0.24
Resolution: 2.80 Å
R/Rfree: 0.21/0.24
X-ray diffraction data for the Crystal Structure of Lysyl-tRNA Synthetase from Cryptosporidium parvum complexed with L-lysine and cladosporin


First author:
Dranow Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.90 Å
R/Rfree: 0.20/0.25
Resolution: 1.90 Å
R/Rfree: 0.20/0.25
X-ray diffraction data for the Crystal structure of a Putative aldehyde dehydrogenase family protein Burkholderia cenocepacia J2315 in complex with partially reduced NADH


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.10 Å
R/Rfree: 0.15/0.19
Resolution: 2.10 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of a glucose-1-phosphate thymidylyltransferase from Burkholderia phymatum bound to 2'-deoxy-thymidine-B-L-rhamnose


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.10 Å
R/Rfree: 0.18/0.23
Resolution: 2.10 Å
R/Rfree: 0.18/0.23
X-ray diffraction data for the Crystal Structure of Ribose-phosphate Pyrophosphokinase from Legionella pneumophila with bound AMP, ADP, and Ribose-5-Phosphate


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.75 Å
R/Rfree: 0.15/0.18
Resolution: 1.75 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of prolyl-tRNA synthetase from Naegleria fowleri in complex with proline and adenosine monophophsphate (AMP)


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.00 Å
R/Rfree: 0.15/0.19
Resolution: 2.00 Å
R/Rfree: 0.15/0.19
X-ray diffraction data for the Crystal structure of N-myristoyl transferase (NMT) G386E mutant from Plasmodium vivax in complex with inhibitor IMP-1002


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.55 Å
R/Rfree: 0.15/0.18
Resolution: 1.55 Å
R/Rfree: 0.15/0.18
X-ray diffraction data for the Crystal structure of N-myristoyl transferase (NMT) G386E mutant from Plasmodium vivax in complex with inhibitor IMP-0366


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.55 Å
R/Rfree: 0.15/0.17
Resolution: 1.55 Å
R/Rfree: 0.15/0.17
X-ray diffraction data for the Crystal Structure of a Probable short-chain type dehydrogenase/reductase (Rv1144) from Mycobacterium tuberculosis with bound NAD


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.20 Å
R/Rfree: 0.12/0.14
Resolution: 1.20 Å
R/Rfree: 0.12/0.14
X-ray diffraction data for the Crystal structure of dihydropteroate synthase from Anaplasma phagocytophilum with bound 6-hydroxymethylpterin-monophosphate


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.00 Å
R/Rfree: 0.16/0.20
Resolution: 2.00 Å
R/Rfree: 0.16/0.20
X-ray diffraction data for the Crystal structure of Glutamate-1-semialdehyde 2,1-aminomutase from Stenotrophomonas maltophilia K279a in complex with PLP


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.40 Å
R/Rfree: 0.13/0.18
Resolution: 1.40 Å
R/Rfree: 0.13/0.18
X-ray diffraction data for the Crystal Structure of Leucine-, isoleucine-, valine-, threonine-, and alanine-binding protein from Brucella ovis, closed conformation


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 1.70 Å
R/Rfree: 0.16/0.19
Resolution: 1.70 Å
R/Rfree: 0.16/0.19
X-ray diffraction data for the Crystal structure of Acetyl-CoA Synthetase in Complex with Adenosine-5'-propylphosphate from Aspergillus fumigatus


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.80 Å
R/Rfree: 0.22/0.25
Resolution: 2.80 Å
R/Rfree: 0.22/0.25
X-ray diffraction data for the Crystal structure of GDP-mannose 4,6-dehydratase from Brucella abortus (strain 2308) in complex with Guanosine-diphosphate-rhamnose


First author:
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
Resolution: 2.35 Å
R/Rfree: 0.17/0.21
Resolution: 2.35 Å
R/Rfree: 0.17/0.21