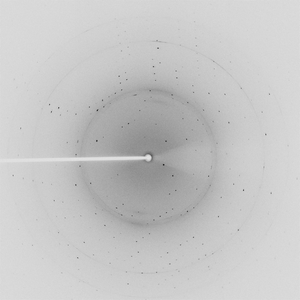
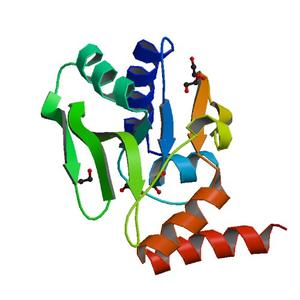
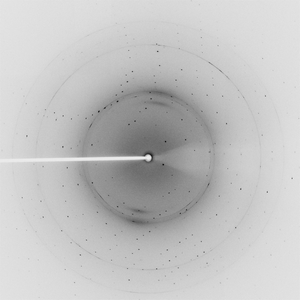
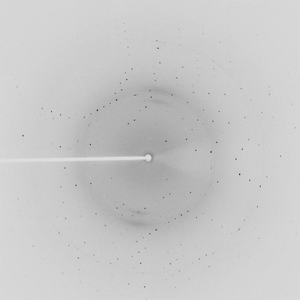
Diffraction project datasets TM1012_2ewr
Project details
| Title |
Crystal structure of a putative nucleotidyltransferase (tm1012) from Thermotoga maritima at 1.60 A resolution |
| Authors |
Joint Center for Structural Genomics (JCSG) |
| Bioentity |
None |
| R / Rfree |
0.15 / 0.18 |
| Unit cell edges [Å] |
32.56 x
36.21 x
39.28
|
| Unit cell angles [°] |
74.1,
86.4,
79.7
|
Dataset 18058_2.#### details
| Number of frames |
240 (1 - 240) |
| Distance [mm] |
250.0 |
| Oscillation width [°] |
1.50 |
| Wavelength [Å] |
0.97934 |
| Experiment Date |
2005-08-04 |
| Equipment |
23-ID-D
at APS (Advanced Photon Source)
|
Dataset 18058_3.#### details
| Number of frames |
240 (1 - 240) |
| Distance [mm] |
250.0 |
| Oscillation width [°] |
1.50 |
| Wavelength [Å] |
0.95373 |
| Experiment Date |
2005-08-04 |
| Equipment |
23-ID-D
at APS (Advanced Photon Source)
|
Dataset 18058_4.#### details
| Number of frames |
240 (1 - 240) |
| Distance [mm] |
250.0 |
| Oscillation width [°] |
1.50 |
| Wavelength [Å] |
0.97934 |
| Experiment Date |
2005-08-04 |
| Equipment |
23-ID-D
at APS (Advanced Photon Source)
|
Dataset 18058_5.#### details
| Number of frames |
107 (1 - 107) |
| Distance [mm] |
250.0 |
| Oscillation width [°] |
1.50 |
| Phi [°] |
0.75 |
| Wavelength [Å] |
0.97934 |
| Experiment Date |
2005-08-04 |
| Equipment |
23-ID-D
at APS (Advanced Photon Source)
|
Dataset 18058_6.#### details
| Number of frames |
150 (1 - 150) |
| Distance [mm] |
250.0 |
| Oscillation width [°] |
1.50 |
| Phi [°] |
158.25 |
| Wavelength [Å] |
0.97934 |
| Experiment Date |
2005-08-04 |
| Equipment |
23-ID-D
at APS (Advanced Photon Source)
|