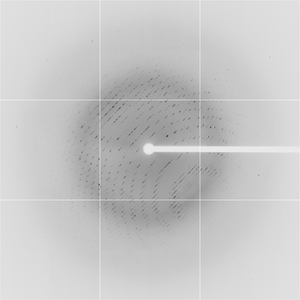
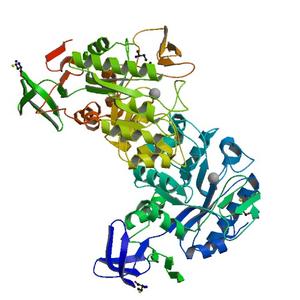
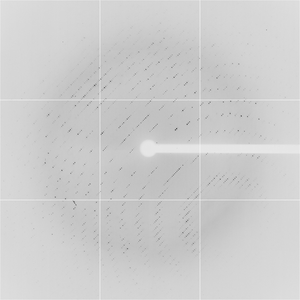
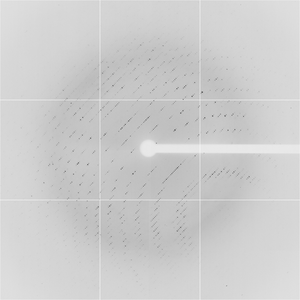
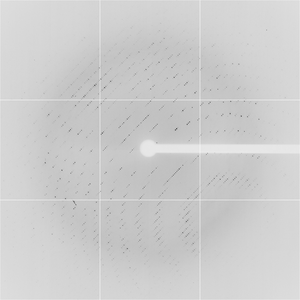
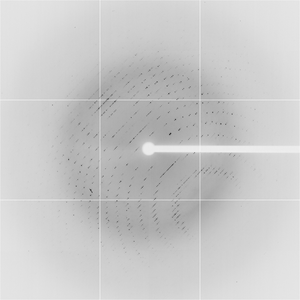
Diffraction project datasets TM0814_1o12
Project details
| Title |
Crystal structure of N-acetylglucosamine-6-phosphate deacetylase (TM0814) from Thermotoga maritima at 2.5 A resolution |
| Authors |
Joint Center for Structural Genomics (JCSG) |
| Bioentity |
None |
| R / Rfree |
0.20 / 0.25 |
| Unit cell edges [Å] |
85.07 x
85.07 x
206.01
|
| Unit cell angles [°] |
90.0,
90.0,
120.0
|
Dataset 21c10p2_1_E1_###.img details
| Number of frames |
30 (1 - 30) |
| Distance [mm] |
250.0 |
| Oscillation width [°] |
1.00 |
| Phi [°] |
90.0 |
| Wavelength [Å] |
0.97903 |
| Equipment |
BL9-2
at SSRL (Stanford Synchrotron Radiation Laboratory)
|
Dataset 21c10p2_2_E1_###.img details
| Number of frames |
133 (1 - 133) |
| Distance [mm] |
375.0 |
| Oscillation width [°] |
1.00 |
| Phi [°] |
90.0 |
| Wavelength [Å] |
0.97903 |
| Equipment |
BL9-2
at SSRL (Stanford Synchrotron Radiation Laboratory)
|
Dataset 21c10p2_2_E2_###.img details
| Number of frames |
120 (1 - 120) |
| Distance [mm] |
375.0 |
| Oscillation width [°] |
1.00 |
| Phi [°] |
90.0 |
| Wavelength [Å] |
0.91837 |
| Equipment |
BL9-2
at SSRL (Stanford Synchrotron Radiation Laboratory)
|
Dataset 21c10p2_2_E3_###.img details
| Number of frames |
120 (1 - 120) |
| Distance [mm] |
375.0 |
| Oscillation width [°] |
1.00 |
| Phi [°] |
90.0 |
| Wavelength [Å] |
0.97874 |
| Equipment |
BL9-2
at SSRL (Stanford Synchrotron Radiation Laboratory)
|
Dataset 21c10p2_3_###.img details
| Number of frames |
180 (1 - 180) |
| Distance [mm] |
290.0 |
| Oscillation width [°] |
0.50 |
| Phi [°] |
90.0 |
| Wavelength [Å] |
0.98397 |
| Equipment |
BL9-2
at SSRL (Stanford Synchrotron Radiation Laboratory)
|
Dataset 21c10p2_4_###.img details
| Number of frames |
180 (1 - 180) |
| Distance [mm] |
450.0 |
| Oscillation width [°] |
1.00 |
| Phi [°] |
90.0 |
| Wavelength [Å] |
0.98397 |
| Equipment |
BL9-2
at SSRL (Stanford Synchrotron Radiation Laboratory)
|