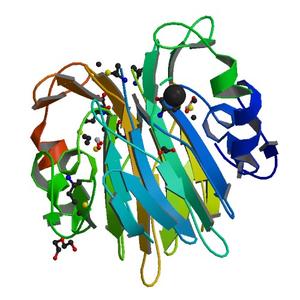
Diffraction project datasets MG6567B_3i7d
Project details
| Title |
Crystal structure of sugar phosphate isomerase from a cupin superfamily SPO2919 from Silicibacter pomeroyi (YP_168127.1) from SILICIBACTER POMEROYI DSS-3 at 2.30 A resolution |
| Authors |
Joint Center for Structural Genomics (JCSG) |
| Bioentity |
None |
| R / Rfree |
0.20 / 0.25 |
| Unit cell edges [Å] |
52.81 x
52.81 x
252.16
|
| Unit cell angles [°] |
90.0,
90.0,
90.0
|
Dataset 105648_1_E1_###.mccd details
| Number of frames |
120 (1 - 120) |
| Distance [mm] |
300.0 |
| Oscillation width [°] |
1.00 |
| Phi [°] |
160.0 |
| Wavelength [Å] |
0.97959 |
| Experiment Date |
2009-01-18 |
| Equipment |
BL9-2
at SSRL (Stanford Synchrotron Radiation Laboratory)
|
Dataset 105648_1_E2_###.mccd details
| Number of frames |
120 (1 - 120) |
| Distance [mm] |
300.0 |
| Oscillation width [°] |
1.00 |
| Phi [°] |
160.0 |
| Wavelength [Å] |
0.91837 |
| Experiment Date |
2009-01-18 |
| Equipment |
BL9-2
at SSRL (Stanford Synchrotron Radiation Laboratory)
|
Dataset 105648_2_###.mccd details
| Number of frames |
120 (1 - 120) |
| Distance [mm] |
300.0 |
| Oscillation width [°] |
1.00 |
| Phi [°] |
160.0 |
| Wavelength [Å] |
0.97941 |
| Experiment Date |
2009-01-18 |
| Equipment |
BL9-2
at SSRL (Stanford Synchrotron Radiation Laboratory)
|