Diffraction project datasets APC102301_3RPZ
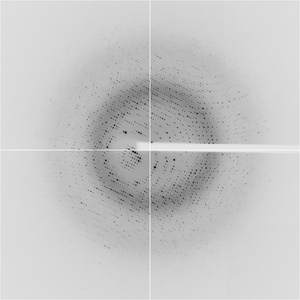
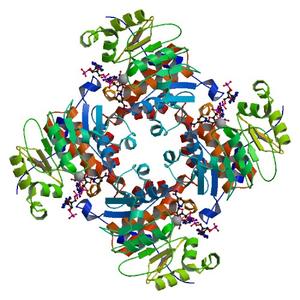
- Method: Molecular Replacement
- Resolution: 1.51 Å
- Space group: I 4 2 2
PDB website for 3RPZ
PDB validation report for 3RPZ
doi:10.18430/M3FK5Q
Project details
| Title | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with NADPH |
| Authors | Shumilin, I.A., Cymborowski, M., Chertihin, O., Jha, K.N., Herr, J.C., Lesley, S.A., Joachimiak, A., Minor, W. |
| Bioentity | None |
| R / Rfree | 0.13 / 0.15 |
| Unit cell edges [Å] | 91.92 x 91.92 x 169.63 |
| Unit cell angles [°] | 90.0, 90.0, 90.0 |
Dataset APC234_Iy-82_1_####.img details

| Number of frames | 180 (1 - 180) |
| Distance [mm] | 140.0 |
| Oscillation width [°] | 1.00 |
| Phi [°] | -116.0 |
| Wavelength [Å] | 0.97918 |
| Equipment | 19-BM at APS (Advanced Photon Source) |