Diffraction project datasets 3md0
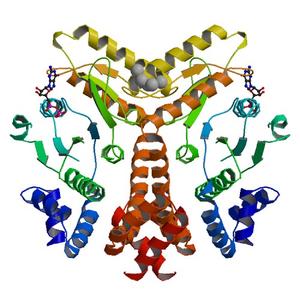
- Method: Molecular Replacement
- Resolution: 2.45 Å
- Space group: C 2 2 21
PDB website for 3MD0
PDB validation report for 3MD0
doi:10.18430/M33MD0
Project details
| Title | Crystal structure of arginine/ornithine transport system ATPase from Mycobacterium tuberculosis bound to GDP (a RAS-like GTPase superfamily protein) |
| Authors | Baugh, L., Phan, I., Begley, D.W., Clifton, M.C., Armour, B., Dranow, D.M., Taylor, B.M., Muruthi, M.M., Abendroth, J., Fairman, J.W., Fox, D., Dieterich, S.H., Staker, B.L., Gardberg, A.S., Choi, R., Hewitt, S.N., Napuli, A.J., Myers, J., Barrett, L.K., Zhang, Y., Ferrell, M., Mundt, E., Thompkins, K., Tran, N., Lyons-Abbott, S., Abramov, A., Sekar, A., Serbzhinskiy, D., Lorimer, D., Buchko, G.W., Stacy, R., Stewart, L.J., Edwards, T.E., Van Voorhis, W.C., Myler, P.J. |
| R / Rfree | 0.21 / 0.25 |
| Unit cell edges [Å] | 65.53 x 187.36 x 66.20 |
| Unit cell angles [°] | 90.0, 90.0, 90.0 |
Dataset 207391d3_y####.img details

| Number of frames | 360 (1 - 360) |
| Distance [mm] | 65.0 |
| Oscillation width [°] | 0.50 |
| Omega [°] | -90.0 |
| Wavelength [Å] | 1.54178 |
| Equipment | HOME_SOURCE at HOME_SOURCE (Home Source) |